Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
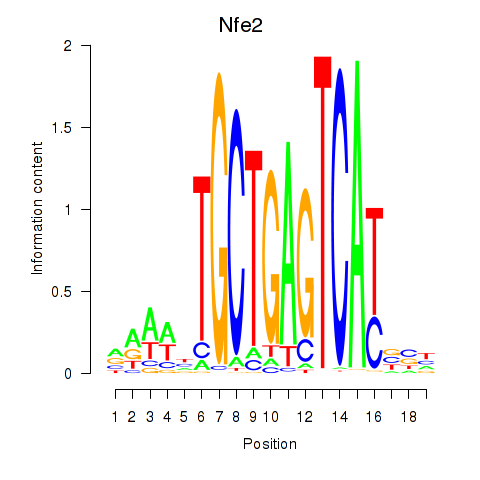
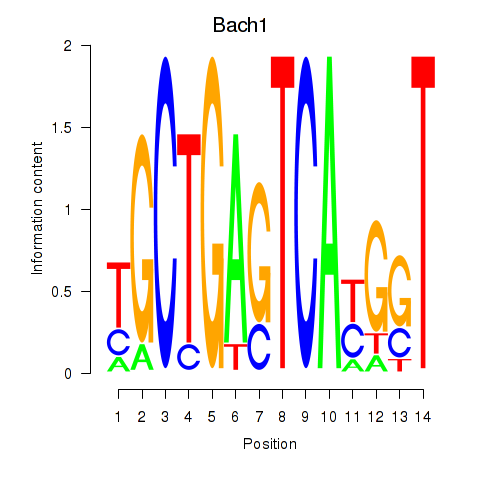
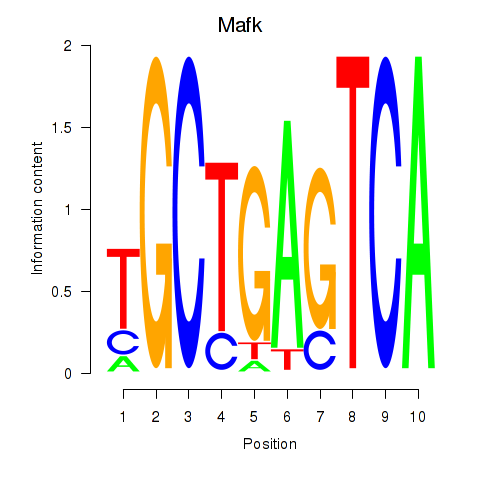
Results for Nfe2_Bach1_Mafk
Z-value: 3.22
Transcription factors associated with Nfe2_Bach1_Mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2
|
ENSMUSG00000058794.13 | nuclear factor, erythroid derived 2 |
|
Bach1
|
ENSMUSG00000025612.6 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
|
Mafk
|
ENSMUSG00000018143.11 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2 | mm39_v1_chr15_-_103161237_103161256 | -0.83 | 8.3e-02 | Click! |
| Mafk | mm39_v1_chr5_+_139777263_139777271 | 0.73 | 1.6e-01 | Click! |
| Bach1 | mm39_v1_chr16_+_87495792_87495872 | -0.47 | 4.2e-01 | Click! |
Activity profile of Nfe2_Bach1_Mafk motif
Sorted Z-values of Nfe2_Bach1_Mafk motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126398165 | 3.87 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr4_-_57956411 | 3.45 |
ENSMUST00000030051.6
|
Txn1
|
thioredoxin 1 |
| chr17_-_45883421 | 3.14 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr4_+_116543045 | 2.90 |
ENSMUST00000129315.8
ENSMUST00000106470.8 |
Prdx1
|
peroxiredoxin 1 |
| chr18_-_43610829 | 2.46 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr14_-_36857202 | 2.08 |
ENSMUST00000165649.4
ENSMUST00000224769.2 |
Ghitm
|
growth hormone inducible transmembrane protein |
| chr6_-_16898440 | 2.02 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr12_+_71021395 | 1.92 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr16_+_20470402 | 1.91 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr4_+_116542741 | 1.80 |
ENSMUST00000135573.8
ENSMUST00000151129.8 |
Prdx1
|
peroxiredoxin 1 |
| chr12_-_40298072 | 1.75 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr14_+_67953547 | 1.71 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr19_-_4092218 | 1.66 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr18_-_35087355 | 1.59 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr11_+_50101717 | 1.58 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr16_+_90017634 | 1.58 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr18_-_10030017 | 1.49 |
ENSMUST00000116669.2
ENSMUST00000092096.14 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr11_-_48762170 | 1.37 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr17_-_45884179 | 1.35 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr14_-_36857083 | 1.35 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr5_-_92496730 | 1.27 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr7_+_127399776 | 1.27 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr19_-_4087907 | 1.25 |
ENSMUST00000237982.2
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr7_+_127399789 | 1.23 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_+_151947444 | 1.20 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr12_+_82303350 | 1.14 |
ENSMUST00000222714.2
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr19_-_32080496 | 1.14 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr14_+_67953687 | 1.11 |
ENSMUST00000150768.8
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr10_-_117215920 | 1.09 |
ENSMUST00000175924.2
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr14_-_79539063 | 1.03 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr5_-_65585720 | 1.01 |
ENSMUST00000131263.3
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr2_+_164790139 | 0.99 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr14_+_8348779 | 0.97 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr1_+_165596961 | 0.96 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr8_+_95710977 | 0.95 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_-_74614654 | 0.94 |
ENSMUST00000102520.9
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr8_-_25581354 | 0.94 |
ENSMUST00000125466.2
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr2_+_150412329 | 0.94 |
ENSMUST00000089200.3
|
Cst7
|
cystatin F (leukocystatin) |
| chr12_+_55445560 | 0.92 |
ENSMUST00000021412.9
|
Psma6
|
proteasome subunit alpha 6 |
| chrX_+_73468140 | 0.90 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr11_+_78069477 | 0.87 |
ENSMUST00000092880.14
ENSMUST00000127587.8 |
Tlcd1
|
TLC domain containing 1 |
| chr16_+_20367327 | 0.85 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr8_-_108315024 | 0.84 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr11_-_30599510 | 0.82 |
ENSMUST00000074613.4
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr1_-_75195127 | 0.81 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr9_-_21150350 | 0.81 |
ENSMUST00000049567.10
ENSMUST00000193982.2 |
Keap1
|
kelch-like ECH-associated protein 1 |
| chr4_-_126215462 | 0.80 |
ENSMUST00000102617.5
|
Adprs
|
ADP-ribosylserine hydrolase |
| chr6_+_129510117 | 0.78 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr7_-_112946481 | 0.78 |
ENSMUST00000117577.8
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr12_+_111505253 | 0.78 |
ENSMUST00000220803.2
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr1_+_85992341 | 0.78 |
ENSMUST00000027432.9
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chrX_-_133483849 | 0.76 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr12_+_111504450 | 0.72 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr3_-_116301684 | 0.69 |
ENSMUST00000153005.2
|
Rtca
|
RNA 3'-terminal phosphate cyclase |
| chrX_-_165992145 | 0.69 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr9_+_107445101 | 0.68 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chrX_+_163052367 | 0.68 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr9_-_21150523 | 0.68 |
ENSMUST00000194542.6
ENSMUST00000216436.2 |
Keap1
|
kelch-like ECH-associated protein 1 |
| chr5_+_136996713 | 0.68 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr4_-_42168603 | 0.67 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr5_-_21990170 | 0.66 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr4_-_42665763 | 0.66 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr12_-_101943134 | 0.66 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr11_+_76836330 | 0.65 |
ENSMUST00000021197.10
|
Blmh
|
bleomycin hydrolase |
| chr11_+_120564185 | 0.65 |
ENSMUST00000135346.8
ENSMUST00000127269.8 ENSMUST00000131727.9 ENSMUST00000149389.8 ENSMUST00000153346.8 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr2_+_163389068 | 0.63 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chrX_+_158155171 | 0.62 |
ENSMUST00000087143.7
|
Eif1ax
|
eukaryotic translation initiation factor 1A, X-linked |
| chr7_+_127400016 | 0.62 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr4_+_155606919 | 0.61 |
ENSMUST00000177094.8
|
Gnb1
|
guanine nucleotide binding protein (G protein), beta 1 |
| chr18_+_77017713 | 0.61 |
ENSMUST00000026487.6
ENSMUST00000136800.2 |
Ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr14_+_79753055 | 0.61 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr11_-_50101592 | 0.61 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr19_-_4087940 | 0.60 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr12_+_111505216 | 0.60 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr7_+_127728712 | 0.59 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr13_-_23894697 | 0.59 |
ENSMUST00000091707.13
ENSMUST00000006787.8 |
Hfe
|
homeostatic iron regulator |
| chr1_+_135945705 | 0.58 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr19_-_8690330 | 0.58 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr18_+_12776358 | 0.57 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr7_+_49408847 | 0.57 |
ENSMUST00000085272.7
ENSMUST00000207895.2 |
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr10_-_121462219 | 0.57 |
ENSMUST00000039810.8
ENSMUST00000218004.2 |
Xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr17_+_35268942 | 0.56 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr1_+_135945798 | 0.56 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr7_-_113875261 | 0.56 |
ENSMUST00000135570.8
|
Psma1
|
proteasome subunit alpha 1 |
| chr8_-_48128164 | 0.56 |
ENSMUST00000080353.3
|
Ing2
|
inhibitor of growth family, member 2 |
| chr17_-_45903410 | 0.55 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_-_19005721 | 0.54 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr15_-_83006289 | 0.54 |
ENSMUST00000166019.2
|
Rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr12_-_84447702 | 0.53 |
ENSMUST00000122194.8
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr11_+_76836545 | 0.53 |
ENSMUST00000125145.8
|
Blmh
|
bleomycin hydrolase |
| chr6_-_67014348 | 0.53 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr2_-_25114660 | 0.53 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr7_+_126843446 | 0.52 |
ENSMUST00000052509.6
|
Zfp771
|
zinc finger protein 771 |
| chr5_+_138115165 | 0.52 |
ENSMUST00000062350.15
ENSMUST00000110961.9 ENSMUST00000080732.10 ENSMUST00000110960.9 ENSMUST00000142185.8 ENSMUST00000136425.2 ENSMUST00000110959.2 |
Zscan21
|
zinc finger and SCAN domain containing 21 |
| chr13_-_55677109 | 0.51 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr14_+_55129950 | 0.51 |
ENSMUST00000140691.8
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr15_-_79718423 | 0.50 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr12_-_84447625 | 0.49 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr14_-_55950545 | 0.49 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr4_-_136613498 | 0.48 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr1_+_127234441 | 0.48 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr14_+_67953584 | 0.48 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr7_-_33929667 | 0.48 |
ENSMUST00000206415.2
|
Gpi1
|
glucose-6-phosphate isomerase 1 |
| chr3_+_28859585 | 0.48 |
ENSMUST00000043867.11
ENSMUST00000194649.2 |
Rpl22l1
|
ribosomal protein L22 like 1 |
| chrX_+_161684221 | 0.47 |
ENSMUST00000101095.9
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr2_+_90884349 | 0.46 |
ENSMUST00000067663.14
ENSMUST00000002171.14 ENSMUST00000111441.10 |
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr11_+_106146966 | 0.46 |
ENSMUST00000021049.9
|
Psmc5
|
protease (prosome, macropain) 26S subunit, ATPase 5 |
| chr18_+_38088597 | 0.46 |
ENSMUST00000070709.9
ENSMUST00000177058.8 ENSMUST00000169360.9 ENSMUST00000163591.9 ENSMUST00000091932.12 |
Rell2
|
RELT-like 2 |
| chr18_-_64649497 | 0.46 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr4_-_58912678 | 0.45 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr5_-_122639840 | 0.45 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr2_+_11710246 | 0.45 |
ENSMUST00000148748.8
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr13_-_8921732 | 0.45 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr8_-_127672590 | 0.45 |
ENSMUST00000179857.3
ENSMUST00000212771.2 |
Tomm20
|
translocase of outer mitochondrial membrane 20 |
| chr10_+_4561974 | 0.45 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr11_+_60619224 | 0.44 |
ENSMUST00000018743.5
|
Mief2
|
mitochondrial elongation factor 2 |
| chr5_+_25452342 | 0.44 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr6_+_138118565 | 0.44 |
ENSMUST00000118091.8
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr19_-_41252370 | 0.43 |
ENSMUST00000237871.2
ENSMUST00000025989.10 |
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr12_+_111504640 | 0.43 |
ENSMUST00000222375.2
ENSMUST00000222388.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr9_+_5298669 | 0.43 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr7_+_120516967 | 0.43 |
ENSMUST00000207481.2
ENSMUST00000106483.4 ENSMUST00000033173.15 |
Polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr11_-_43638790 | 0.43 |
ENSMUST00000048578.3
ENSMUST00000109278.8 |
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr11_+_60428788 | 0.42 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr14_-_52257113 | 0.42 |
ENSMUST00000166169.4
ENSMUST00000226605.2 |
Zfp219
|
zinc finger protein 219 |
| chr10_+_128641663 | 0.42 |
ENSMUST00000218511.2
|
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr10_+_115979787 | 0.42 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr2_+_153491363 | 0.41 |
ENSMUST00000072997.10
ENSMUST00000109773.8 ENSMUST00000109774.9 ENSMUST00000081628.13 ENSMUST00000103151.8 ENSMUST00000056495.14 ENSMUST00000088976.12 ENSMUST00000109772.8 ENSMUST00000103150.10 |
Dnmt3b
|
DNA methyltransferase 3B |
| chr1_-_88437617 | 0.41 |
ENSMUST00000067625.9
|
Glrp1
|
glutamine repeat protein 1 |
| chr11_-_120520954 | 0.40 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr2_+_90884612 | 0.40 |
ENSMUST00000185715.7
|
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr6_+_51500881 | 0.40 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr6_-_67512768 | 0.40 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr12_+_51737775 | 0.40 |
ENSMUST00000218820.2
ENSMUST00000021338.10 |
Ap4s1
|
adaptor-related protein complex AP-4, sigma 1 |
| chr9_-_21150100 | 0.39 |
ENSMUST00000164812.8
|
Keap1
|
kelch-like ECH-associated protein 1 |
| chr4_-_43558386 | 0.39 |
ENSMUST00000130353.2
|
Tln1
|
talin 1 |
| chr4_-_140501507 | 0.39 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr9_+_108566513 | 0.39 |
ENSMUST00000192344.2
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr9_-_60594742 | 0.38 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr11_-_120521382 | 0.38 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr17_+_25059079 | 0.38 |
ENSMUST00000164251.8
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr19_+_8816663 | 0.38 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_-_85335193 | 0.38 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr14_-_20443676 | 0.38 |
ENSMUST00000061444.5
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr16_-_22258469 | 0.37 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr10_+_128626772 | 0.37 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr7_-_104002534 | 0.37 |
ENSMUST00000130139.3
ENSMUST00000059037.15 |
Trim12c
|
tripartite motif-containing 12C |
| chr18_-_64649620 | 0.36 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr3_+_95836558 | 0.36 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr2_+_118731860 | 0.36 |
ENSMUST00000036578.7
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr9_+_54858388 | 0.36 |
ENSMUST00000171900.2
|
Psma4
|
proteasome subunit alpha 4 |
| chr2_-_25162347 | 0.35 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr11_-_78402931 | 0.35 |
ENSMUST00000052566.8
|
Tmem199
|
transmembrane protein 199 |
| chr2_-_20948230 | 0.34 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr1_-_30988772 | 0.34 |
ENSMUST00000238874.2
ENSMUST00000027232.15 ENSMUST00000076587.6 ENSMUST00000233506.2 |
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr9_-_103243039 | 0.34 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr3_+_88243562 | 0.34 |
ENSMUST00000001451.11
|
Smg5
|
Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chrX_-_37341274 | 0.34 |
ENSMUST00000089056.10
ENSMUST00000089054.11 ENSMUST00000066498.8 |
Tmem255a
|
transmembrane protein 255A |
| chr10_-_43777712 | 0.34 |
ENSMUST00000020012.7
|
Qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr15_+_80139371 | 0.33 |
ENSMUST00000109605.5
ENSMUST00000229828.2 |
Atf4
|
activating transcription factor 4 |
| chr6_-_120334400 | 0.33 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr16_-_18161746 | 0.33 |
ENSMUST00000231372.2
ENSMUST00000130752.2 ENSMUST00000231605.2 ENSMUST00000115628.10 |
Tango2
|
transport and golgi organization 2 |
| chr4_-_109333866 | 0.33 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr6_-_67014191 | 0.31 |
ENSMUST00000204282.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr11_-_50216426 | 0.31 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr7_-_33929708 | 0.31 |
ENSMUST00000038027.6
|
Gpi1
|
glucose-6-phosphate isomerase 1 |
| chr8_-_70389147 | 0.30 |
ENSMUST00000212277.2
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr7_-_113875342 | 0.30 |
ENSMUST00000033008.10
|
Psma1
|
proteasome subunit alpha 1 |
| chr11_-_106146868 | 0.29 |
ENSMUST00000021048.7
|
Ftsj3
|
FtsJ RNA methyltransferase homolog 3 (E. coli) |
| chr9_+_64024429 | 0.29 |
ENSMUST00000034969.14
|
Lctl
|
lactase-like |
| chr7_+_127160751 | 0.28 |
ENSMUST00000190278.3
|
Tmem265
|
transmembrane protein 265 |
| chr11_-_109364424 | 0.28 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_-_4799345 | 0.28 |
ENSMUST00000053079.13
ENSMUST00000109910.9 |
Nf2
|
neurofibromin 2 |
| chr11_-_116080361 | 0.27 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr2_+_163837423 | 0.27 |
ENSMUST00000131288.2
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr6_-_86770504 | 0.27 |
ENSMUST00000204441.3
ENSMUST00000204398.2 ENSMUST00000001187.15 |
Anxa4
|
annexin A4 |
| chr7_-_126074222 | 0.27 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr17_-_56440817 | 0.27 |
ENSMUST00000167545.3
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr8_+_111782951 | 0.26 |
ENSMUST00000210390.2
|
Exosc6
|
exosome component 6 |
| chr19_+_56414114 | 0.26 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr12_+_86999366 | 0.26 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr10_+_127894816 | 0.26 |
ENSMUST00000052798.14
|
Ptges3
|
prostaglandin E synthase 3 |
| chr8_-_68427217 | 0.25 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_+_7272514 | 0.25 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr15_+_25774070 | 0.25 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr11_+_78237492 | 0.24 |
ENSMUST00000100755.4
|
Unc119
|
unc-119 lipid binding chaperone |
| chr2_+_29968596 | 0.24 |
ENSMUST00000045246.8
|
Pkn3
|
protein kinase N3 |
| chr5_-_108022900 | 0.24 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr5_-_137784943 | 0.24 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme |
| chrX_-_153999333 | 0.24 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr11_+_78403019 | 0.24 |
ENSMUST00000001127.11
|
Poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr5_+_147797258 | 0.24 |
ENSMUST00000031654.10
|
Pomp
|
proteasome maturation protein |
| chr17_-_45903188 | 0.24 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr9_-_20085353 | 0.24 |
ENSMUST00000215984.3
|
Olfr870
|
olfactory receptor 870 |
| chr11_-_120930193 | 0.23 |
ENSMUST00000026159.6
|
Cd7
|
CD7 antigen |
| chrX_-_72974357 | 0.23 |
ENSMUST00000155597.2
ENSMUST00000114379.8 |
Renbp
|
renin binding protein |
| chr1_+_58069090 | 0.23 |
ENSMUST00000001027.7
|
Aox1
|
aldehyde oxidase 1 |
| chr2_+_155617251 | 0.22 |
ENSMUST00000029141.6
|
Mmp24
|
matrix metallopeptidase 24 |
| chr19_+_8906916 | 0.22 |
ENSMUST00000096241.6
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr5_+_114268425 | 0.22 |
ENSMUST00000031587.13
|
Ung
|
uracil DNA glycosylase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2_Bach1_Mafk
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:1903659 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.6 | 1.9 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.4 | 1.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.4 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 1.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 3.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.3 | 1.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 1.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 1.3 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.3 | 3.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 1.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.3 | 0.8 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.2 | 0.7 | GO:1904437 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.2 | 0.7 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 0.6 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.2 | 6.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 3.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 0.6 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.2 | 0.6 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 | 1.0 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 0.8 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 2.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.4 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.1 | 0.8 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.5 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.4 | GO:0071449 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.9 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.8 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.4 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.2 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.8 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.4 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.9 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.3 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.6 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.4 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.8 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.4 | GO:0061727 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.6 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.9 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 1.0 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.0 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 2.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 1.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 1.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.1 | GO:0070256 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 1.0 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.7 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 1.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0015881 | creatine transport(GO:0015881) |
| 0.0 | 0.1 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.7 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.0 | GO:2001274 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 3.5 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.4 | 4.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 4.0 | GO:0031597 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.2 | 0.6 | GO:0044753 | amphisome(GO:0044753) |
| 0.2 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 0.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.2 | 0.5 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.2 | 4.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.9 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:1990589 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.8 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.1 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.8 | 3.3 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.6 | 4.5 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 3.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.4 | 3.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 2.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 4.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 1.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 0.9 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.3 | 1.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 1.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 1.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.6 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.2 | 1.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 0.8 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 1.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 0.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 4.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.4 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 2.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.4 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.2 | GO:0043546 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 3.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.2 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 3.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 4.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 3.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 10.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |