Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
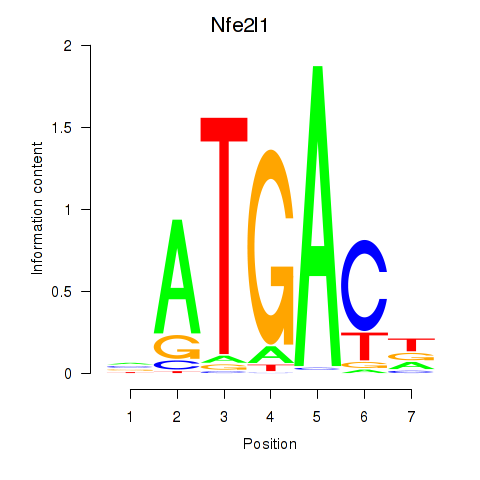
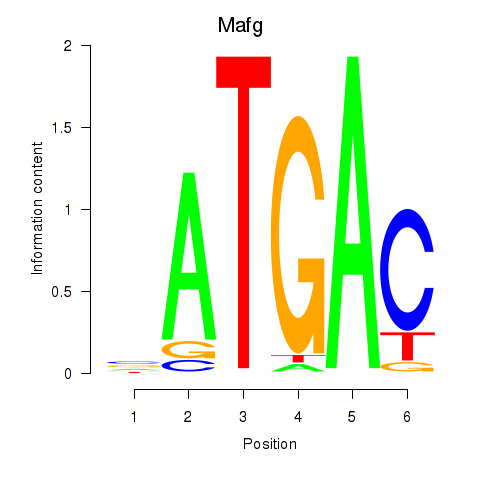
Results for Nfe2l1_Mafg
Z-value: 0.23
Transcription factors associated with Nfe2l1_Mafg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l1
|
ENSMUSG00000038615.18 | nuclear factor, erythroid derived 2,-like 1 |
|
Mafg
|
ENSMUSG00000051510.14 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2l1 | mm39_v1_chr11_-_96720738_96720794 | 0.86 | 6.0e-02 | Click! |
| Mafg | mm39_v1_chr11_-_120524362_120524431 | 0.71 | 1.8e-01 | Click! |
Activity profile of Nfe2l1_Mafg motif
Sorted Z-values of Nfe2l1_Mafg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_43610829 | 0.16 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr12_-_31684588 | 0.15 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr5_+_31070739 | 0.14 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr2_-_64853083 | 0.14 |
ENSMUST00000028252.14
|
Grb14
|
growth factor receptor bound protein 14 |
| chr3_+_121220146 | 0.14 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr7_-_16651107 | 0.11 |
ENSMUST00000173139.2
|
Calm3
|
calmodulin 3 |
| chr4_+_102427247 | 0.09 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr6_-_24528012 | 0.09 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr19_-_10079091 | 0.09 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr8_-_61407760 | 0.09 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr1_+_87998487 | 0.09 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr10_-_13744676 | 0.08 |
ENSMUST00000019942.6
ENSMUST00000162610.8 |
Aig1
|
androgen-induced 1 |
| chr2_-_25162347 | 0.08 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr19_+_8719033 | 0.08 |
ENSMUST00000176314.8
ENSMUST00000073430.14 ENSMUST00000175901.8 |
Stx5a
|
syntaxin 5A |
| chr10_+_80765900 | 0.08 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr6_+_17749169 | 0.07 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr9_+_122942280 | 0.07 |
ENSMUST00000026891.5
ENSMUST00000215377.2 |
Exosc7
|
exosome component 7 |
| chr7_-_79989965 | 0.07 |
ENSMUST00000127997.2
ENSMUST00000032748.15 |
Unc45a
|
unc-45 myosin chaperone A |
| chr9_+_108437485 | 0.07 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr7_+_127172701 | 0.06 |
ENSMUST00000121004.3
|
Phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr12_-_55033130 | 0.06 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr18_-_80194682 | 0.06 |
ENSMUST00000066743.11
|
Adnp2
|
ADNP homeobox 2 |
| chr2_+_130119077 | 0.06 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr9_-_35122261 | 0.06 |
ENSMUST00000043805.15
ENSMUST00000142595.8 ENSMUST00000127996.8 |
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr12_+_55445560 | 0.06 |
ENSMUST00000021412.9
|
Psma6
|
proteasome subunit alpha 6 |
| chr2_+_120439858 | 0.06 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr12_-_56392646 | 0.05 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr7_+_82516491 | 0.05 |
ENSMUST00000082237.7
|
Mex3b
|
mex3 RNA binding family member B |
| chr11_-_116164928 | 0.05 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr11_-_83421333 | 0.05 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr5_+_145060489 | 0.05 |
ENSMUST00000136074.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr2_+_25162487 | 0.05 |
ENSMUST00000028341.11
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr12_+_84408742 | 0.05 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr12_+_84408803 | 0.05 |
ENSMUST00000110278.8
ENSMUST00000145522.2 |
Coq6
|
coenzyme Q6 monooxygenase |
| chr8_-_68427217 | 0.05 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_97689263 | 0.05 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_+_48691175 | 0.05 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr17_-_71305003 | 0.05 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_-_129484070 | 0.04 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr14_-_31362835 | 0.04 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr4_+_126915104 | 0.04 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chrX_-_153999440 | 0.04 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr2_+_132689640 | 0.04 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr12_+_17398421 | 0.04 |
ENSMUST00000046011.12
|
Nol10
|
nucleolar protein 10 |
| chr10_+_82821304 | 0.04 |
ENSMUST00000040110.8
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr13_-_55677109 | 0.04 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr7_+_101546059 | 0.04 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr2_+_112092271 | 0.04 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr5_-_107873883 | 0.04 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr11_-_116165024 | 0.04 |
ENSMUST00000021133.16
|
Srp68
|
signal recognition particle 68 |
| chr6_+_11926757 | 0.04 |
ENSMUST00000133776.2
|
Phf14
|
PHD finger protein 14 |
| chr1_+_127234441 | 0.04 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr8_+_85786684 | 0.04 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr3_-_104960437 | 0.04 |
ENSMUST00000077548.12
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chrX_+_55500170 | 0.04 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr15_+_8138805 | 0.04 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chr15_+_82031382 | 0.04 |
ENSMUST00000023100.8
ENSMUST00000229336.2 |
Srebf2
|
sterol regulatory element binding factor 2 |
| chr5_-_65248927 | 0.03 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr11_+_93935021 | 0.03 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr9_-_66702638 | 0.03 |
ENSMUST00000113730.3
|
Aph1b
|
aph1 homolog B, gamma secretase subunit |
| chr9_+_20914211 | 0.03 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr11_+_83553400 | 0.03 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr11_+_93935066 | 0.03 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr7_-_27749453 | 0.03 |
ENSMUST00000140053.3
ENSMUST00000032824.10 |
Psmc4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr2_-_120439981 | 0.03 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr5_-_138185438 | 0.03 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr5_+_24679154 | 0.03 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr3_+_88523730 | 0.03 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr16_+_35803794 | 0.03 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr3_-_104960264 | 0.03 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr13_-_38819012 | 0.03 |
ENSMUST00000224902.2
ENSMUST00000224115.2 ENSMUST00000035899.8 |
Bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr10_+_58207229 | 0.03 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_-_34495329 | 0.03 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr11_+_60428788 | 0.03 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr13_-_55676334 | 0.03 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr14_-_70945434 | 0.03 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr19_+_8907206 | 0.03 |
ENSMUST00000224272.2
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr8_-_76133212 | 0.03 |
ENSMUST00000212864.2
|
Gm10358
|
predicted gene 10358 |
| chr11_+_51510555 | 0.03 |
ENSMUST00000127405.2
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr2_-_121101822 | 0.03 |
ENSMUST00000110647.8
ENSMUST00000110648.8 |
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr8_+_124100588 | 0.03 |
ENSMUST00000211932.2
ENSMUST00000212569.2 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr17_+_87590308 | 0.03 |
ENSMUST00000041110.12
ENSMUST00000125875.8 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr2_-_112198366 | 0.03 |
ENSMUST00000028551.4
|
Emc4
|
ER membrane protein complex subunit 4 |
| chr17_-_56933872 | 0.03 |
ENSMUST00000047226.10
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr2_-_52225763 | 0.03 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr11_+_86574811 | 0.02 |
ENSMUST00000108022.8
ENSMUST00000108021.2 |
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr5_-_65548723 | 0.02 |
ENSMUST00000120094.5
ENSMUST00000127874.6 ENSMUST00000057885.13 |
Rpl9
|
ribosomal protein L9 |
| chr3_+_89970088 | 0.02 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr5_+_65548840 | 0.02 |
ENSMUST00000122026.6
ENSMUST00000031101.10 ENSMUST00000200374.2 |
Lias
|
lipoic acid synthetase |
| chr2_-_52225146 | 0.02 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr11_+_93935156 | 0.02 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr1_+_87983189 | 0.02 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_87983099 | 0.02 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr14_+_31363004 | 0.02 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr17_+_71511642 | 0.02 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr13_-_101831020 | 0.02 |
ENSMUST00000185795.2
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr1_+_87332638 | 0.02 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr2_-_121101794 | 0.02 |
ENSMUST00000131245.2
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr14_-_72840373 | 0.02 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr5_-_138185686 | 0.02 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr8_+_125624615 | 0.02 |
ENSMUST00000034467.7
|
Sprtn
|
SprT-like N-terminal domain |
| chr9_+_111011388 | 0.02 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_-_79884920 | 0.02 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr11_-_85125889 | 0.02 |
ENSMUST00000018625.10
|
Appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr14_-_31362909 | 0.02 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr17_-_34250616 | 0.02 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr1_+_82817170 | 0.02 |
ENSMUST00000189220.7
ENSMUST00000113444.8 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr2_-_7400690 | 0.02 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr11_+_93934940 | 0.02 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr2_-_105832353 | 0.02 |
ENSMUST00000155811.2
|
Dnajc24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr11_+_97253221 | 0.02 |
ENSMUST00000238729.2
ENSMUST00000045540.4 |
Socs7
|
suppressor of cytokine signaling 7 |
| chr17_-_35077089 | 0.02 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr19_-_5781503 | 0.02 |
ENSMUST00000162976.2
|
Znrd2
|
zinc ribbon domain containing 2 |
| chr9_+_108765701 | 0.02 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr15_-_102097466 | 0.02 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr11_-_109363406 | 0.02 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr7_+_127441119 | 0.02 |
ENSMUST00000121705.8
|
Stx4a
|
syntaxin 4A (placental) |
| chr14_+_29700319 | 0.02 |
ENSMUST00000224797.2
|
Actr8
|
ARP8 actin-related protein 8 |
| chr4_-_129534403 | 0.02 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr9_-_79885063 | 0.02 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr14_+_79718604 | 0.02 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chrX_+_70600481 | 0.02 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chrX_-_161671421 | 0.02 |
ENSMUST00000033723.4
|
Syap1
|
synapse associated protein 1 |
| chr2_-_120439826 | 0.02 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr17_+_36132567 | 0.02 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr15_-_102097387 | 0.02 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr10_+_111000613 | 0.02 |
ENSMUST00000105275.9
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr1_-_182929025 | 0.02 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr13_-_54759086 | 0.02 |
ENSMUST00000049575.8
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr3_+_88536749 | 0.01 |
ENSMUST00000176316.8
ENSMUST00000176879.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr17_-_26004298 | 0.01 |
ENSMUST00000150324.8
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr5_-_65248962 | 0.01 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr1_-_128256048 | 0.01 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr16_-_17711950 | 0.01 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr14_+_58063668 | 0.01 |
ENSMUST00000022538.5
|
Mrpl57
|
mitochondrial ribosomal protein L57 |
| chr19_+_38825120 | 0.01 |
ENSMUST00000235558.2
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12 |
| chr11_+_72498029 | 0.01 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr6_+_88701810 | 0.01 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr9_+_53678801 | 0.01 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr8_+_124100492 | 0.01 |
ENSMUST00000212571.2
ENSMUST00000212470.2 ENSMUST00000108840.4 ENSMUST00000057934.10 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr2_-_120439764 | 0.01 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr7_+_127440924 | 0.01 |
ENSMUST00000033075.14
|
Stx4a
|
syntaxin 4A (placental) |
| chr10_-_128727542 | 0.01 |
ENSMUST00000026408.7
|
Gdf11
|
growth differentiation factor 11 |
| chr6_+_88701578 | 0.01 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr10_-_13744523 | 0.01 |
ENSMUST00000105534.10
|
Aig1
|
androgen-induced 1 |
| chr9_-_83028654 | 0.01 |
ENSMUST00000161796.9
ENSMUST00000162246.9 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr6_-_114898739 | 0.01 |
ENSMUST00000032459.14
|
Vgll4
|
vestigial like family member 4 |
| chr19_-_5416339 | 0.01 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr5_-_139812162 | 0.01 |
ENSMUST00000182839.2
|
Gm26938
|
predicted gene, 26938 |
| chr18_-_3281089 | 0.01 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr11_-_115824290 | 0.01 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr11_-_51891575 | 0.01 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr11_-_50216426 | 0.01 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr3_+_88536805 | 0.01 |
ENSMUST00000175745.2
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_12245527 | 0.01 |
ENSMUST00000073507.3
|
Olfr235
|
olfactory receptor 235 |
| chr3_+_88523440 | 0.01 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr14_+_50630215 | 0.01 |
ENSMUST00000058965.4
|
Olfr736
|
olfactory receptor 736 |
| chr14_+_29700294 | 0.01 |
ENSMUST00000016115.6
|
Actr8
|
ARP8 actin-related protein 8 |
| chr1_-_36312482 | 0.01 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr7_+_30475819 | 0.01 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr8_+_95710977 | 0.01 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr6_+_41107047 | 0.01 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr19_+_12186932 | 0.01 |
ENSMUST00000072316.5
|
Olfr1431
|
olfactory receptor 1431 |
| chr11_+_100978103 | 0.01 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr15_-_42540363 | 0.01 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr11_-_70128587 | 0.01 |
ENSMUST00000108576.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr2_-_7400780 | 0.01 |
ENSMUST00000002176.13
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr4_-_129534752 | 0.01 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr17_-_46956920 | 0.01 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chr9_-_22041894 | 0.01 |
ENSMUST00000115315.3
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr17_-_34822649 | 0.01 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr7_-_109559593 | 0.01 |
ENSMUST00000106722.2
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr18_-_3280999 | 0.01 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr1_-_163552693 | 0.01 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr2_-_86938729 | 0.01 |
ENSMUST00000099862.2
|
Olfr259
|
olfactory receptor 259 |
| chr9_-_123778334 | 0.01 |
ENSMUST00000071404.5
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr11_-_109188892 | 0.01 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr7_-_115637970 | 0.01 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr14_-_50322136 | 0.01 |
ENSMUST00000072370.3
|
Olfr726
|
olfactory receptor 726 |
| chr12_-_85017586 | 0.01 |
ENSMUST00000165886.2
ENSMUST00000167448.8 ENSMUST00000043169.14 |
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr11_-_83540175 | 0.01 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr5_+_138185747 | 0.01 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr1_-_133681419 | 0.01 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_-_109188947 | 0.01 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr17_+_37777099 | 0.01 |
ENSMUST00000031086.5
|
Olfr109
|
olfactory receptor 109 |
| chr3_-_144514386 | 0.01 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr8_-_70389147 | 0.01 |
ENSMUST00000212277.2
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr12_+_38833501 | 0.01 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr12_+_38833454 | 0.01 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr1_-_84818223 | 0.01 |
ENSMUST00000186465.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr2_+_87248394 | 0.01 |
ENSMUST00000054974.2
|
Olfr1123
|
olfactory receptor 1123 |
| chr15_-_101801351 | 0.01 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr2_+_87201878 | 0.01 |
ENSMUST00000062555.2
|
Olfr1121
|
olfactory receptor 1121 |
| chr2_+_119572770 | 0.00 |
ENSMUST00000028758.8
|
Itpka
|
inositol 1,4,5-trisphosphate 3-kinase A |
| chr8_-_68363564 | 0.00 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_-_86437119 | 0.00 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr13_+_65300040 | 0.00 |
ENSMUST00000058907.4
|
Olfr466
|
olfactory receptor 466 |
| chr17_+_93506590 | 0.00 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr16_+_17712061 | 0.00 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr2_-_89127612 | 0.00 |
ENSMUST00000099787.2
|
Olfr1230
|
olfactory receptor 1230 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l1_Mafg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.0 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |