Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
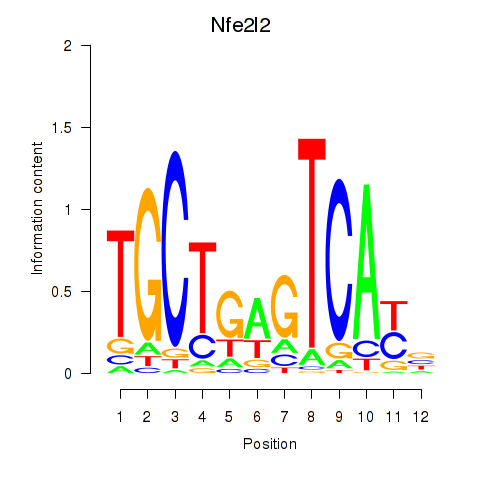
Results for Nfe2l2
Z-value: 1.96
Transcription factors associated with Nfe2l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l2
|
ENSMUSG00000015839.7 | nuclear factor, erythroid derived 2, like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2l2 | mm39_v1_chr2_-_75534985_75535023 | -0.81 | 9.5e-02 | Click! |
Activity profile of Nfe2l2 motif
Sorted Z-values of Nfe2l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_36857202 | 1.65 |
ENSMUST00000165649.4
ENSMUST00000224769.2 |
Ghitm
|
growth hormone inducible transmembrane protein |
| chr17_-_45883421 | 1.50 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr4_+_116543045 | 1.29 |
ENSMUST00000129315.8
ENSMUST00000106470.8 |
Prdx1
|
peroxiredoxin 1 |
| chr7_-_126398165 | 1.23 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr6_-_16898440 | 1.14 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr19_-_4092218 | 1.12 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr2_+_150412329 | 1.12 |
ENSMUST00000089200.3
|
Cst7
|
cystatin F (leukocystatin) |
| chr11_-_74614654 | 1.08 |
ENSMUST00000102520.9
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr12_+_71021395 | 1.07 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr18_-_43610829 | 1.04 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr14_-_36857083 | 1.01 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr4_-_57956411 | 0.98 |
ENSMUST00000030051.6
|
Txn1
|
thioredoxin 1 |
| chr16_+_20470402 | 0.96 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr4_-_126215462 | 0.96 |
ENSMUST00000102617.5
|
Adprs
|
ADP-ribosylserine hydrolase |
| chr2_+_153491363 | 0.90 |
ENSMUST00000072997.10
ENSMUST00000109773.8 ENSMUST00000109774.9 ENSMUST00000081628.13 ENSMUST00000103151.8 ENSMUST00000056495.14 ENSMUST00000088976.12 ENSMUST00000109772.8 ENSMUST00000103150.10 |
Dnmt3b
|
DNA methyltransferase 3B |
| chr14_+_67953547 | 0.89 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr7_-_113875261 | 0.82 |
ENSMUST00000135570.8
|
Psma1
|
proteasome subunit alpha 1 |
| chr7_-_80475722 | 0.80 |
ENSMUST00000205304.2
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr5_+_45677571 | 0.80 |
ENSMUST00000156481.8
ENSMUST00000119579.3 ENSMUST00000118833.3 |
Med28
|
mediator complex subunit 28 |
| chr3_+_108164242 | 0.78 |
ENSMUST00000090569.10
|
Psma5
|
proteasome subunit alpha 5 |
| chr17_-_71305003 | 0.74 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_+_151947444 | 0.72 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr19_-_4087907 | 0.71 |
ENSMUST00000237982.2
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr11_+_120123727 | 0.71 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr12_-_40298072 | 0.69 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr5_+_137756407 | 0.67 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chrX_+_158155171 | 0.65 |
ENSMUST00000087143.7
|
Eif1ax
|
eukaryotic translation initiation factor 1A, X-linked |
| chr12_+_55445560 | 0.65 |
ENSMUST00000021412.9
|
Psma6
|
proteasome subunit alpha 6 |
| chr4_+_116542741 | 0.65 |
ENSMUST00000135573.8
ENSMUST00000151129.8 |
Prdx1
|
peroxiredoxin 1 |
| chr2_+_112092271 | 0.65 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr5_-_45607485 | 0.64 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr5_-_92496730 | 0.63 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr5_-_45607463 | 0.63 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr6_-_68713748 | 0.62 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr19_-_8690330 | 0.62 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr10_-_117215920 | 0.62 |
ENSMUST00000175924.2
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr12_-_85335193 | 0.59 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr14_+_67953687 | 0.58 |
ENSMUST00000150768.8
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr11_+_50101717 | 0.58 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr18_-_35087355 | 0.57 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr11_-_59700820 | 0.55 |
ENSMUST00000047706.3
ENSMUST00000102697.10 |
Flcn
|
folliculin |
| chr16_+_90017634 | 0.54 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr11_+_60428788 | 0.53 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr19_-_32080496 | 0.53 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr5_-_45607554 | 0.52 |
ENSMUST00000015950.12
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr12_-_83609217 | 0.51 |
ENSMUST00000222448.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr12_+_111505253 | 0.51 |
ENSMUST00000220803.2
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr3_-_146487102 | 0.49 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr10_+_17672004 | 0.48 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chrX_+_73468140 | 0.48 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr17_-_45884179 | 0.47 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr1_-_75195127 | 0.47 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr1_+_135945798 | 0.47 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr2_+_120439858 | 0.46 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr5_+_100946493 | 0.46 |
ENSMUST00000016977.15
ENSMUST00000112898.8 ENSMUST00000112901.2 |
Mrps18c
|
mitochondrial ribosomal protein S18C |
| chr1_+_135945705 | 0.46 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr8_-_108315024 | 0.44 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr16_+_16962464 | 0.44 |
ENSMUST00000231726.2
|
Ydjc
|
YdjC homolog (bacterial) |
| chr9_-_21150350 | 0.44 |
ENSMUST00000049567.10
ENSMUST00000193982.2 |
Keap1
|
kelch-like ECH-associated protein 1 |
| chr7_-_113875342 | 0.43 |
ENSMUST00000033008.10
|
Psma1
|
proteasome subunit alpha 1 |
| chr11_+_78069477 | 0.43 |
ENSMUST00000092880.14
ENSMUST00000127587.8 |
Tlcd1
|
TLC domain containing 1 |
| chr14_-_20443676 | 0.43 |
ENSMUST00000061444.5
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr15_+_80139371 | 0.42 |
ENSMUST00000109605.5
ENSMUST00000229828.2 |
Atf4
|
activating transcription factor 4 |
| chr11_+_76836330 | 0.41 |
ENSMUST00000021197.10
|
Blmh
|
bleomycin hydrolase |
| chr2_+_90884349 | 0.41 |
ENSMUST00000067663.14
ENSMUST00000002171.14 ENSMUST00000111441.10 |
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr11_+_120564185 | 0.40 |
ENSMUST00000135346.8
ENSMUST00000127269.8 ENSMUST00000131727.9 ENSMUST00000149389.8 ENSMUST00000153346.8 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr11_+_76836545 | 0.40 |
ENSMUST00000125145.8
|
Blmh
|
bleomycin hydrolase |
| chr15_-_79718423 | 0.40 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr5_-_138185438 | 0.39 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr7_+_127399789 | 0.39 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr19_-_4889314 | 0.39 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr2_-_25162347 | 0.38 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr9_-_21150523 | 0.37 |
ENSMUST00000194542.6
ENSMUST00000216436.2 |
Keap1
|
kelch-like ECH-associated protein 1 |
| chr7_+_127399776 | 0.36 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr3_-_89867869 | 0.36 |
ENSMUST00000069805.14
|
Atp8b2
|
ATPase, class I, type 8B, member 2 |
| chrX_-_133483849 | 0.35 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr13_-_55677109 | 0.35 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr16_-_18161746 | 0.35 |
ENSMUST00000231372.2
ENSMUST00000130752.2 ENSMUST00000231605.2 ENSMUST00000115628.10 |
Tango2
|
transport and golgi organization 2 |
| chr12_+_111505216 | 0.34 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr2_+_90884612 | 0.34 |
ENSMUST00000185715.7
|
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr5_-_21990170 | 0.34 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr7_-_30810422 | 0.34 |
ENSMUST00000039435.15
|
Hpn
|
hepsin |
| chr2_-_120439981 | 0.33 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr2_-_166885414 | 0.33 |
ENSMUST00000067584.7
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr7_+_140438468 | 0.33 |
ENSMUST00000209766.2
|
Ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr1_+_85992341 | 0.33 |
ENSMUST00000027432.9
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr11_-_55075855 | 0.33 |
ENSMUST00000039305.6
|
Slc36a2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr1_-_155022501 | 0.31 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr8_+_95710977 | 0.31 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr1_+_157353696 | 0.28 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr11_-_30599510 | 0.28 |
ENSMUST00000074613.4
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr8_+_106052970 | 0.28 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr2_+_164790139 | 0.27 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr19_-_4889284 | 0.27 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr19_-_4087940 | 0.27 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr16_+_20367327 | 0.26 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chrX_-_37341274 | 0.26 |
ENSMUST00000089056.10
ENSMUST00000089054.11 ENSMUST00000066498.8 |
Tmem255a
|
transmembrane protein 255A |
| chr3_+_28859585 | 0.26 |
ENSMUST00000043867.11
ENSMUST00000194649.2 |
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr6_+_48818567 | 0.26 |
ENSMUST00000203639.3
|
Tmem176a
|
transmembrane protein 176A |
| chr12_+_111504450 | 0.25 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr10_+_127894816 | 0.25 |
ENSMUST00000052798.14
|
Ptges3
|
prostaglandin E synthase 3 |
| chr14_+_67953584 | 0.24 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr9_-_44162762 | 0.24 |
ENSMUST00000034618.6
|
Pdzd3
|
PDZ domain containing 3 |
| chr16_-_50151350 | 0.24 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr5_-_138185686 | 0.24 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr7_+_127511681 | 0.24 |
ENSMUST00000033070.9
|
Kat8
|
K(lysine) acetyltransferase 8 |
| chr7_+_127400016 | 0.24 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr9_-_21150100 | 0.24 |
ENSMUST00000164812.8
|
Keap1
|
kelch-like ECH-associated protein 1 |
| chr14_-_55950545 | 0.23 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr1_+_165596961 | 0.23 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr2_+_25162487 | 0.23 |
ENSMUST00000028341.11
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr7_+_101875019 | 0.23 |
ENSMUST00000142873.8
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr2_+_155617251 | 0.22 |
ENSMUST00000029141.6
|
Mmp24
|
matrix metallopeptidase 24 |
| chr10_+_115979787 | 0.22 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr4_-_109333866 | 0.21 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr15_+_78294154 | 0.21 |
ENSMUST00000229739.2
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr7_+_127160751 | 0.21 |
ENSMUST00000190278.3
|
Tmem265
|
transmembrane protein 265 |
| chr11_-_120930193 | 0.20 |
ENSMUST00000026159.6
|
Cd7
|
CD7 antigen |
| chr7_-_104002534 | 0.20 |
ENSMUST00000130139.3
ENSMUST00000059037.15 |
Trim12c
|
tripartite motif-containing 12C |
| chr4_+_102278715 | 0.20 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_120439764 | 0.20 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr7_+_127728712 | 0.19 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr2_-_120439826 | 0.19 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr5_+_20112500 | 0.19 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_+_97253221 | 0.19 |
ENSMUST00000238729.2
ENSMUST00000045540.4 |
Socs7
|
suppressor of cytokine signaling 7 |
| chrX_-_165992145 | 0.19 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr11_-_50216426 | 0.19 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr18_+_35731735 | 0.18 |
ENSMUST00000236868.2
|
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr11_+_3845221 | 0.17 |
ENSMUST00000109996.8
ENSMUST00000055931.5 |
Dusp18
|
dual specificity phosphatase 18 |
| chr4_-_140501507 | 0.17 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chrX_+_163052367 | 0.17 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr13_-_23894828 | 0.16 |
ENSMUST00000091706.14
|
Hfe
|
homeostatic iron regulator |
| chr7_+_18915136 | 0.16 |
ENSMUST00000144054.8
ENSMUST00000141718.8 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr11_-_120520954 | 0.15 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr2_+_11710246 | 0.15 |
ENSMUST00000148748.8
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr10_+_127894843 | 0.15 |
ENSMUST00000084771.3
|
Ptges3
|
prostaglandin E synthase 3 |
| chr12_+_111504640 | 0.15 |
ENSMUST00000222375.2
ENSMUST00000222388.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr11_-_120521382 | 0.15 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr9_+_108566513 | 0.15 |
ENSMUST00000192344.2
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr4_-_133601990 | 0.14 |
ENSMUST00000168974.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr4_+_12140263 | 0.14 |
ENSMUST00000050069.9
ENSMUST00000069128.8 |
Rbm12b1
|
RNA binding motif protein 12 B1 |
| chr13_+_49735938 | 0.14 |
ENSMUST00000221170.2
|
Omd
|
osteomodulin |
| chr11_+_107370375 | 0.14 |
ENSMUST00000106750.5
|
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr5_+_138185747 | 0.13 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr19_+_8906916 | 0.13 |
ENSMUST00000096241.6
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr11_+_107370303 | 0.13 |
ENSMUST00000021063.13
ENSMUST00000106752.10 |
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr12_-_17050332 | 0.13 |
ENSMUST00000134938.8
ENSMUST00000137960.2 |
Pqlc3
|
PQ loop repeat containing |
| chr5_-_100946334 | 0.12 |
ENSMUST00000133845.8
|
Helq
|
helicase, POLQ-like |
| chr7_+_18915086 | 0.12 |
ENSMUST00000120595.8
ENSMUST00000048502.10 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr9_+_32135540 | 0.12 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_-_100946434 | 0.11 |
ENSMUST00000044684.14
|
Helq
|
helicase, POLQ-like |
| chr8_+_70275079 | 0.11 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr5_-_112400375 | 0.11 |
ENSMUST00000112385.8
|
Cryba4
|
crystallin, beta A4 |
| chr1_-_121260298 | 0.11 |
ENSMUST00000071064.13
|
Insig2
|
insulin induced gene 2 |
| chr6_+_117988399 | 0.10 |
ENSMUST00000164960.4
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr11_+_69523754 | 0.10 |
ENSMUST00000018909.4
|
Fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr12_+_16860931 | 0.10 |
ENSMUST00000020908.9
|
E2f6
|
E2F transcription factor 6 |
| chr14_-_122035225 | 0.10 |
ENSMUST00000100299.11
|
Dock9
|
dedicator of cytokinesis 9 |
| chr9_-_20085353 | 0.10 |
ENSMUST00000215984.3
|
Olfr870
|
olfactory receptor 870 |
| chr2_+_166647426 | 0.10 |
ENSMUST00000099078.10
|
Arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr7_+_127399848 | 0.10 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr11_-_50101592 | 0.09 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr7_+_45289391 | 0.09 |
ENSMUST00000148532.4
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr7_-_44646645 | 0.09 |
ENSMUST00000207342.2
|
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr11_-_69696428 | 0.08 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr18_-_3281089 | 0.08 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr1_+_58069090 | 0.08 |
ENSMUST00000001027.7
|
Aox1
|
aldehyde oxidase 1 |
| chr4_-_118667141 | 0.07 |
ENSMUST00000084313.5
ENSMUST00000219094.2 |
Olfr1335
|
olfactory receptor 1335 |
| chr11_-_120328458 | 0.07 |
ENSMUST00000044271.15
ENSMUST00000103017.4 |
Nploc4
|
NPL4 homolog, ubiquitin recognition factor |
| chr18_-_39652468 | 0.07 |
ENSMUST00000237944.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr16_-_16962256 | 0.06 |
ENSMUST00000115711.10
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr4_+_101365144 | 0.06 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr10_+_75404482 | 0.06 |
ENSMUST00000134503.8
ENSMUST00000125770.8 ENSMUST00000128886.8 ENSMUST00000151212.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr7_+_44498415 | 0.06 |
ENSMUST00000107885.8
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr1_-_120001752 | 0.06 |
ENSMUST00000056089.8
|
Tmem37
|
transmembrane protein 37 |
| chr19_+_8816663 | 0.06 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr9_+_65121944 | 0.05 |
ENSMUST00000069000.9
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr17_-_43003135 | 0.05 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr4_+_117109204 | 0.05 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr2_-_44817173 | 0.05 |
ENSMUST00000130991.8
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr5_+_20112704 | 0.05 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_-_141687987 | 0.05 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr5_+_30218112 | 0.05 |
ENSMUST00000026845.12
ENSMUST00000199183.5 ENSMUST00000195978.5 |
Il6
|
interleukin 6 |
| chr18_-_3280999 | 0.04 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr7_+_108312527 | 0.04 |
ENSMUST00000209620.4
ENSMUST00000074730.4 |
Olfr512
|
olfactory receptor 512 |
| chr11_+_4207557 | 0.04 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr18_+_35731658 | 0.04 |
ENSMUST00000041314.17
ENSMUST00000236666.2 ENSMUST00000236020.2 ENSMUST00000235400.2 |
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr16_-_32688640 | 0.04 |
ENSMUST00000089684.10
ENSMUST00000040986.15 ENSMUST00000115105.9 |
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr12_-_113760187 | 0.04 |
ENSMUST00000192911.2
ENSMUST00000103455.3 |
Ighv2-6-8
|
immunoglobulin heavy variable 2-6-8 |
| chr14_-_52257452 | 0.03 |
ENSMUST00000228162.2
|
Zfp219
|
zinc finger protein 219 |
| chr14_+_53921052 | 0.03 |
ENSMUST00000103663.6
|
Trav4-4-dv10
|
T cell receptor alpha variable 4-4-DV10 |
| chr18_+_20380397 | 0.03 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr3_-_144514386 | 0.03 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr9_-_44437694 | 0.03 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr12_-_113649535 | 0.03 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr4_+_130297132 | 0.03 |
ENSMUST00000105993.4
|
Nkain1
|
Na+/K+ transporting ATPase interacting 1 |
| chr17_+_37756371 | 0.03 |
ENSMUST00000078207.4
ENSMUST00000218675.2 |
Olfr108
|
olfactory receptor 108 |
| chr19_-_8906686 | 0.03 |
ENSMUST00000096242.5
|
Rom1
|
rod outer segment membrane protein 1 |
| chrX_-_35755483 | 0.03 |
ENSMUST00000169499.2
|
Gm14569
|
predicted gene 14569 |
| chr11_+_78068931 | 0.03 |
ENSMUST00000147819.8
|
Tlcd1
|
TLC domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1903660 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.3 | 1.0 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.3 | 0.9 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.2 | 0.8 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.2 | 0.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 0.6 | GO:1901874 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.2 | 1.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 1.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.2 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.6 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 1.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.4 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 2.5 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 0.3 | GO:0036233 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 0.1 | 1.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.8 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.2 | GO:1904440 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.7 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0070814 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 0.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.2 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.4 | 2.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 3.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 2.0 | GO:0031597 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.0 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.6 | 1.8 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.3 | 2.0 | GO:0002135 | CTP binding(GO:0002135) |
| 0.3 | 1.1 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 1.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 3.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.5 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.7 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 6.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 2.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |