Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nfia
Z-value: 0.97
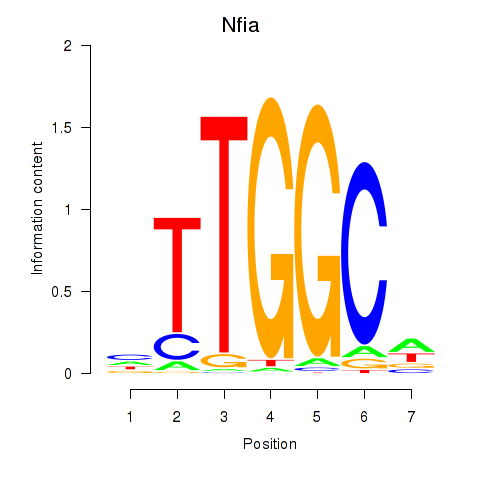
Transcription factors associated with Nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfia
|
ENSMUSG00000028565.19 | nuclear factor I/A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfia | mm39_v1_chr4_+_97665992_97666071 | -0.36 | 5.6e-01 | Click! |
Activity profile of Nfia motif
Sorted Z-values of Nfia motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_21938258 | 1.51 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr13_-_23806530 | 1.26 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr13_-_23945189 | 0.97 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr13_-_23735822 | 0.88 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr13_-_23746543 | 0.76 |
ENSMUST00000105107.2
|
H3c6
|
H3 clustered histone 6 |
| chr13_-_21937997 | 0.74 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr13_+_21906214 | 0.74 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr13_+_21919225 | 0.66 |
ENSMUST00000087714.6
|
H4c11
|
H4 clustered histone 11 |
| chr10_-_128245501 | 0.62 |
ENSMUST00000172348.8
ENSMUST00000166608.8 ENSMUST00000164199.8 ENSMUST00000171370.2 ENSMUST00000026439.14 |
Nabp2
|
nucleic acid binding protein 2 |
| chr7_+_43000765 | 0.53 |
ENSMUST00000012798.14
ENSMUST00000122423.8 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr7_+_43000838 | 0.44 |
ENSMUST00000206299.2
ENSMUST00000121494.2 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr17_+_7292967 | 0.41 |
ENSMUST00000097422.6
|
Gm1604b
|
predicted gene 1604b |
| chr1_+_107456731 | 0.41 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr13_+_22017906 | 0.40 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr7_+_46700349 | 0.38 |
ENSMUST00000010451.8
|
Tmem86a
|
transmembrane protein 86A |
| chr8_+_66419809 | 0.38 |
ENSMUST00000072482.13
|
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr12_-_45120895 | 0.38 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr9_-_48747232 | 0.37 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr17_-_26727437 | 0.35 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr2_+_121978156 | 0.33 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chrX_-_71318353 | 0.31 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr5_+_52521133 | 0.31 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr7_+_26895206 | 0.30 |
ENSMUST00000179391.8
ENSMUST00000108379.8 |
BC024978
|
cDNA sequence BC024978 |
| chr7_-_43956326 | 0.30 |
ENSMUST00000004587.11
|
Clec11a
|
C-type lectin domain family 11, member a |
| chr14_+_20732804 | 0.27 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr2_-_5680801 | 0.27 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr9_-_48747474 | 0.27 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr17_+_84819260 | 0.26 |
ENSMUST00000047206.7
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr2_-_32277773 | 0.25 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr8_-_106723026 | 0.24 |
ENSMUST00000227363.2
ENSMUST00000081998.13 |
Dpep2
|
dipeptidase 2 |
| chr15_-_76501525 | 0.24 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr8_+_69333143 | 0.23 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr7_+_79939747 | 0.23 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr11_+_106916430 | 0.23 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chr15_-_101268036 | 0.23 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr4_-_4138432 | 0.23 |
ENSMUST00000070375.8
|
Penk
|
preproenkephalin |
| chr18_-_62313019 | 0.22 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr7_-_26895561 | 0.22 |
ENSMUST00000122202.8
ENSMUST00000080356.10 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr11_+_69471219 | 0.22 |
ENSMUST00000108657.4
|
Trp53
|
transformation related protein 53 |
| chr15_+_81350538 | 0.22 |
ENSMUST00000230219.2
|
Rbx1
|
ring-box 1 |
| chr4_+_47091909 | 0.21 |
ENSMUST00000045041.12
ENSMUST00000107744.2 |
Galnt12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr7_-_25358406 | 0.21 |
ENSMUST00000071329.8
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr10_+_128061699 | 0.21 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr11_-_50183129 | 0.21 |
ENSMUST00000059458.5
|
Maml1
|
mastermind like transcriptional coactivator 1 |
| chr11_+_69214971 | 0.21 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr13_-_22225527 | 0.21 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr16_+_51851917 | 0.20 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr11_+_77239098 | 0.20 |
ENSMUST00000181283.3
|
Ssh2
|
slingshot protein phosphatase 2 |
| chr11_+_116324913 | 0.20 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr6_+_49013601 | 0.20 |
ENSMUST00000204260.2
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr3_-_115923098 | 0.20 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr7_+_26958150 | 0.20 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr10_-_30679289 | 0.19 |
ENSMUST00000215725.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr2_-_170248421 | 0.19 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr5_+_135093056 | 0.19 |
ENSMUST00000071263.7
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr8_+_71917554 | 0.19 |
ENSMUST00000048914.8
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chr8_-_72763462 | 0.19 |
ENSMUST00000003574.5
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr7_-_19411866 | 0.19 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr7_-_19415301 | 0.18 |
ENSMUST00000150569.9
ENSMUST00000127648.4 ENSMUST00000003071.10 |
Gm44805
Apoc4
|
predicted gene 44805 apolipoprotein C-IV |
| chr19_-_24454720 | 0.18 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr9_-_49479791 | 0.18 |
ENSMUST00000194252.6
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr7_+_25386418 | 0.17 |
ENSMUST00000002678.10
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr8_+_120729459 | 0.17 |
ENSMUST00000108972.4
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr12_-_44257109 | 0.17 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr7_-_24459736 | 0.17 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr11_+_77107006 | 0.17 |
ENSMUST00000156488.8
ENSMUST00000037912.12 |
Ssh2
|
slingshot protein phosphatase 2 |
| chr17_+_24946793 | 0.16 |
ENSMUST00000170239.9
|
Rpl3l
|
ribosomal protein L3-like |
| chr10_-_31485180 | 0.16 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chr4_-_155947819 | 0.16 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr7_-_26895141 | 0.16 |
ENSMUST00000163311.9
ENSMUST00000126211.2 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr6_+_49013517 | 0.16 |
ENSMUST00000031840.10
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr16_+_17269845 | 0.16 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr11_+_33997114 | 0.16 |
ENSMUST00000109329.9
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr16_+_10363222 | 0.15 |
ENSMUST00000155633.8
ENSMUST00000066345.15 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr8_+_13455080 | 0.15 |
ENSMUST00000033827.8
ENSMUST00000209909.2 |
Grk1
|
G protein-coupled receptor kinase 1 |
| chr7_+_140658101 | 0.15 |
ENSMUST00000106039.9
|
Pkp3
|
plakophilin 3 |
| chr1_-_44258112 | 0.15 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr12_-_70394074 | 0.15 |
ENSMUST00000223160.2
ENSMUST00000222316.2 ENSMUST00000167755.3 ENSMUST00000110520.10 ENSMUST00000110522.10 ENSMUST00000221041.2 ENSMUST00000222603.3 |
Trim9
|
tripartite motif-containing 9 |
| chr17_+_46991906 | 0.15 |
ENSMUST00000233430.2
|
Mea1
|
male enhanced antigen 1 |
| chrX_-_47297436 | 0.15 |
ENSMUST00000037960.11
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr17_+_46991972 | 0.15 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr5_+_34817704 | 0.15 |
ENSMUST00000074651.11
ENSMUST00000001112.14 |
Grk4
|
G protein-coupled receptor kinase 4 |
| chr16_+_20492861 | 0.15 |
ENSMUST00000151679.3
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr1_+_88022776 | 0.15 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr13_-_100881117 | 0.15 |
ENSMUST00000078573.5
ENSMUST00000109333.8 |
Mrps36
|
mitochondrial ribosomal protein S36 |
| chr7_+_27878894 | 0.15 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr16_+_10363203 | 0.15 |
ENSMUST00000115824.10
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr7_+_142030744 | 0.14 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr6_+_138117295 | 0.14 |
ENSMUST00000008684.11
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr15_-_5273659 | 0.14 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr5_-_113957362 | 0.14 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr10_-_128759331 | 0.14 |
ENSMUST00000153731.8
ENSMUST00000026405.10 |
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr19_+_45036220 | 0.14 |
ENSMUST00000084493.8
|
Sfxn3
|
sideroflexin 3 |
| chr8_+_122457302 | 0.14 |
ENSMUST00000026357.12
|
Jph3
|
junctophilin 3 |
| chr19_+_8828132 | 0.14 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr12_-_85421467 | 0.14 |
ENSMUST00000040766.9
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr15_-_5273645 | 0.14 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr2_-_73284262 | 0.14 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr14_-_64654397 | 0.14 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr16_+_51851588 | 0.14 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr8_-_33374282 | 0.13 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chrX_+_158491589 | 0.13 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_+_62737936 | 0.13 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr7_-_101513300 | 0.13 |
ENSMUST00000106981.8
|
Folr1
|
folate receptor 1 (adult) |
| chr1_+_165591315 | 0.13 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr8_+_13034245 | 0.13 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr19_+_8817883 | 0.13 |
ENSMUST00000086058.13
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_-_106469938 | 0.13 |
ENSMUST00000103070.3
|
Tex2
|
testis expressed gene 2 |
| chr6_+_135174975 | 0.13 |
ENSMUST00000111915.8
ENSMUST00000111916.2 |
Fam234b
|
family with sequence similarity 234, member B |
| chr11_-_69304501 | 0.13 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr19_+_45036037 | 0.13 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr2_+_164611812 | 0.13 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr14_-_33700719 | 0.13 |
ENSMUST00000166737.2
|
Zfp488
|
zinc finger protein 488 |
| chr19_-_7194912 | 0.13 |
ENSMUST00000039758.6
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chrX_+_162691978 | 0.13 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr5_+_64969679 | 0.13 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr18_+_75500600 | 0.13 |
ENSMUST00000026999.10
|
Smad7
|
SMAD family member 7 |
| chr9_-_103569984 | 0.12 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr11_+_69214789 | 0.12 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr10_-_62363217 | 0.12 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr8_-_123338157 | 0.12 |
ENSMUST00000015171.11
|
Galns
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr12_+_84820024 | 0.12 |
ENSMUST00000021667.7
ENSMUST00000222449.2 ENSMUST00000222982.2 |
Isca2
|
iron-sulfur cluster assembly 2 |
| chr13_-_98951890 | 0.12 |
ENSMUST00000040340.16
ENSMUST00000179563.8 ENSMUST00000109403.2 |
Fcho2
|
FCH domain only 2 |
| chr17_-_57529827 | 0.12 |
ENSMUST00000177425.2
|
C3
|
complement component 3 |
| chrX_-_47297746 | 0.12 |
ENSMUST00000088935.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chrX_+_72760183 | 0.12 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr7_+_27173187 | 0.12 |
ENSMUST00000068641.8
|
Sertad3
|
SERTA domain containing 3 |
| chr6_+_138117519 | 0.12 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr11_-_100288566 | 0.12 |
ENSMUST00000001592.15
ENSMUST00000107403.2 |
Jup
|
junction plakoglobin |
| chr17_-_47813201 | 0.12 |
ENSMUST00000233174.2
ENSMUST00000233121.2 ENSMUST00000067103.4 |
Taf8
|
TATA-box binding protein associated factor 8 |
| chr8_+_95710977 | 0.12 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr6_-_52203146 | 0.12 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr11_-_78313043 | 0.12 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr15_-_42540363 | 0.11 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr13_+_109397184 | 0.11 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_-_109502243 | 0.11 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr8_+_56393488 | 0.11 |
ENSMUST00000000275.10
|
Glra3
|
glycine receptor, alpha 3 subunit |
| chr10_+_125802084 | 0.11 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr11_+_100960838 | 0.11 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr14_-_64654592 | 0.11 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr11_-_46203047 | 0.11 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_102916637 | 0.11 |
ENSMUST00000112852.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr16_+_20492564 | 0.11 |
ENSMUST00000141034.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr9_+_44684450 | 0.11 |
ENSMUST00000238800.2
ENSMUST00000147559.8 |
Ift46
|
intraflagellar transport 46 |
| chr19_-_10859087 | 0.11 |
ENSMUST00000144681.2
|
Tmem109
|
transmembrane protein 109 |
| chr14_-_55909314 | 0.11 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr5_+_150119860 | 0.11 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr7_+_112806672 | 0.11 |
ENSMUST00000047321.9
ENSMUST00000210074.2 ENSMUST00000210238.2 |
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr6_-_124733441 | 0.11 |
ENSMUST00000088357.12
|
Atn1
|
atrophin 1 |
| chr16_+_20492014 | 0.11 |
ENSMUST00000154950.8
ENSMUST00000115461.8 ENSMUST00000136713.5 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr3_-_88366159 | 0.11 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr11_+_75400889 | 0.11 |
ENSMUST00000042972.7
|
Rilp
|
Rab interacting lysosomal protein |
| chr9_-_87613301 | 0.11 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr7_+_3648264 | 0.11 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr10_-_49664839 | 0.11 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr1_+_59952131 | 0.10 |
ENSMUST00000036540.12
|
Fam117b
|
family with sequence similarity 117, member B |
| chr17_+_34824827 | 0.10 |
ENSMUST00000037489.15
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr4_+_137196080 | 0.10 |
ENSMUST00000030547.15
ENSMUST00000171332.2 |
Hspg2
|
perlecan (heparan sulfate proteoglycan 2) |
| chr16_+_27208660 | 0.10 |
ENSMUST00000143823.2
|
Ccdc50
|
coiled-coil domain containing 50 |
| chr16_-_4831349 | 0.10 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chr11_+_77353218 | 0.10 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6 |
| chr11_+_62737887 | 0.10 |
ENSMUST00000036085.11
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr9_+_89791943 | 0.10 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr8_+_71207326 | 0.10 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr9_+_39932760 | 0.10 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr4_-_133695264 | 0.10 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_+_48045143 | 0.10 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr5_+_102629240 | 0.10 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr7_+_27879650 | 0.10 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr17_-_7215682 | 0.10 |
ENSMUST00000024572.10
|
Rsph3b
|
radial spoke 3B homolog (Chlamydomonas) |
| chr18_-_78166595 | 0.10 |
ENSMUST00000091813.12
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr14_-_73622638 | 0.10 |
ENSMUST00000228637.2
ENSMUST00000022704.9 |
Itm2b
|
integral membrane protein 2B |
| chr12_-_44256843 | 0.10 |
ENSMUST00000220421.2
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr15_-_103161237 | 0.10 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr7_-_80053063 | 0.10 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr19_+_16933471 | 0.10 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr8_+_95712151 | 0.10 |
ENSMUST00000212799.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_-_26717686 | 0.10 |
ENSMUST00000162702.8
ENSMUST00000040398.14 ENSMUST00000066560.13 |
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr17_+_29537798 | 0.09 |
ENSMUST00000114701.10
|
Pi16
|
peptidase inhibitor 16 |
| chr4_-_135221810 | 0.09 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr8_+_125302843 | 0.09 |
ENSMUST00000093033.6
ENSMUST00000133086.2 |
Capn9
|
calpain 9 |
| chr13_+_49658249 | 0.09 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr11_-_94492688 | 0.09 |
ENSMUST00000103164.4
|
Acsf2
|
acyl-CoA synthetase family member 2 |
| chr10_-_57408585 | 0.09 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr11_+_69804714 | 0.09 |
ENSMUST00000072581.9
ENSMUST00000116358.8 |
Gps2
|
G protein pathway suppressor 2 |
| chr11_+_69214883 | 0.09 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr2_-_129139125 | 0.09 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr19_+_23736205 | 0.09 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr10_+_67373691 | 0.09 |
ENSMUST00000048289.14
ENSMUST00000130933.2 ENSMUST00000105438.9 ENSMUST00000146986.2 |
Egr2
|
early growth response 2 |
| chr3_+_146156220 | 0.09 |
ENSMUST00000061937.13
ENSMUST00000029840.4 |
Ctbs
|
chitobiase |
| chr7_-_126101555 | 0.09 |
ENSMUST00000167759.8
|
Atxn2l
|
ataxin 2-like |
| chr5_+_123153072 | 0.09 |
ENSMUST00000051016.5
ENSMUST00000121652.8 |
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr16_+_84571249 | 0.09 |
ENSMUST00000098407.3
|
Jam2
|
junction adhesion molecule 2 |
| chr4_+_141095415 | 0.09 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr10_+_75242745 | 0.09 |
ENSMUST00000039925.8
|
Upb1
|
ureidopropionase, beta |
| chr7_-_126101484 | 0.09 |
ENSMUST00000166682.9
|
Atxn2l
|
ataxin 2-like |
| chr8_+_84682136 | 0.09 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr7_+_44667377 | 0.09 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr15_+_81350497 | 0.09 |
ENSMUST00000023036.7
|
Rbx1
|
ring-box 1 |
| chr11_+_78006391 | 0.09 |
ENSMUST00000155571.2
|
Fam222b
|
family with sequence similarity 222, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfia
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.1 | 0.3 | GO:1904438 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 0.3 | GO:1990773 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.1 | 0.6 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:0042376 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.2 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.2 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.2 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.1 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.3 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:1900625 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.6 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0072249 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0033371 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.1 | GO:1900108 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.0 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.0 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0097466 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.0 | GO:2000836 | positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.3 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.2 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.2 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.1 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.0 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0030151 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |