Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
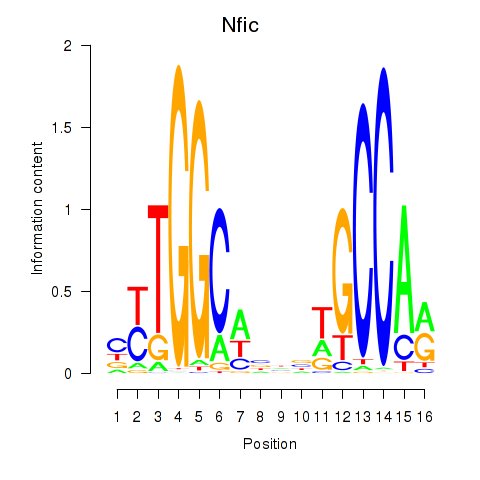
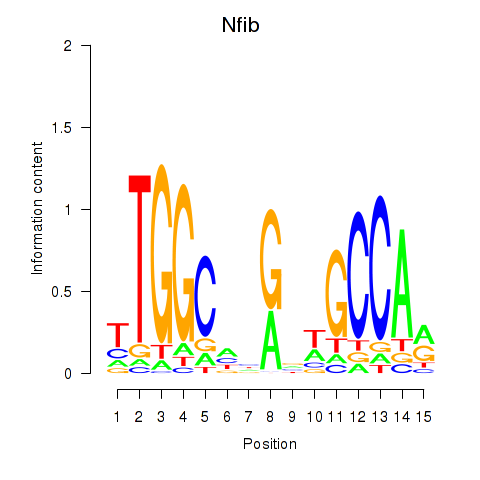
Results for Nfic_Nfib
Z-value: 0.70
Transcription factors associated with Nfic_Nfib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfic
|
ENSMUSG00000055053.19 | nuclear factor I/C |
|
Nfib
|
ENSMUSG00000008575.18 | nuclear factor I/B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfib | mm39_v1_chr4_-_82423511_82423595 | -0.97 | 6.1e-03 | Click! |
| Nfic | mm39_v1_chr10_-_81266800_81266872 | -0.92 | 2.6e-02 | Click! |
Activity profile of Nfic_Nfib motif
Sorted Z-values of Nfic_Nfib motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_96941637 | 0.44 |
ENSMUST00000168565.2
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr16_+_20536415 | 0.34 |
ENSMUST00000021405.8
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr19_-_42741148 | 0.34 |
ENSMUST00000236659.2
ENSMUST00000076505.4 |
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr11_+_78215026 | 0.33 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr4_-_123644091 | 0.32 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr9_+_88209250 | 0.32 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr14_+_30856687 | 0.31 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr9_-_100388857 | 0.31 |
ENSMUST00000112874.4
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chrX_-_11994625 | 0.31 |
ENSMUST00000145872.8
|
Bcor
|
BCL6 interacting corepressor |
| chr5_-_77262968 | 0.30 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr1_+_172328768 | 0.29 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr3_+_121220146 | 0.29 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr7_+_100970910 | 0.29 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr19_+_4644365 | 0.28 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr16_+_32480040 | 0.27 |
ENSMUST00000238806.2
ENSMUST00000238856.2 |
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr8_+_95710977 | 0.26 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr3_-_94566107 | 0.25 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr14_-_55828511 | 0.25 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr18_+_37939442 | 0.25 |
ENSMUST00000076807.7
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr10_-_80513927 | 0.25 |
ENSMUST00000199949.2
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr12_+_3941728 | 0.24 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr15_+_44291470 | 0.24 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr3_+_94600863 | 0.24 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr4_+_102617495 | 0.23 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr11_+_94520567 | 0.23 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr17_-_28736483 | 0.22 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr3_+_51324022 | 0.22 |
ENSMUST00000192419.6
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr12_-_112640378 | 0.22 |
ENSMUST00000130342.2
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr6_+_135339929 | 0.22 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_96941420 | 0.22 |
ENSMUST00000090020.13
|
Osbpl7
|
oxysterol binding protein-like 7 |
| chr12_-_112640626 | 0.21 |
ENSMUST00000001780.10
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr5_+_139239247 | 0.21 |
ENSMUST00000138508.8
ENSMUST00000110878.2 |
Get4
|
golgi to ER traffic protein 4 |
| chr11_+_109376432 | 0.21 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr3_+_88523730 | 0.20 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chrX_+_108138965 | 0.20 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr14_+_51181956 | 0.20 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr9_-_106315518 | 0.20 |
ENSMUST00000024031.13
ENSMUST00000190972.3 |
Acy1
|
aminoacylase 1 |
| chrX_+_20529137 | 0.20 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr17_+_29538157 | 0.20 |
ENSMUST00000114699.9
ENSMUST00000155348.3 ENSMUST00000234618.2 |
Pi16
|
peptidase inhibitor 16 |
| chr15_+_98006346 | 0.20 |
ENSMUST00000051226.8
|
Pfkm
|
phosphofructokinase, muscle |
| chr4_+_102617332 | 0.20 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr9_+_59563838 | 0.19 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chr9_-_56068282 | 0.19 |
ENSMUST00000034876.10
|
Tspan3
|
tetraspanin 3 |
| chr10_+_128173603 | 0.18 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr17_+_47908025 | 0.18 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr3_-_89008406 | 0.18 |
ENSMUST00000200659.2
|
Gm43738
|
predicted gene 43738 |
| chr10_+_81012465 | 0.18 |
ENSMUST00000047864.11
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr4_+_139350152 | 0.18 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr3_+_94840352 | 0.18 |
ENSMUST00000090839.12
|
Selenbp1
|
selenium binding protein 1 |
| chr9_+_59563872 | 0.18 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr2_+_36120438 | 0.18 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr14_+_62529924 | 0.17 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr7_+_100143250 | 0.17 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_-_115915315 | 0.17 |
ENSMUST00000016703.8
|
H3f3b
|
H3.3 histone B |
| chr9_+_69361348 | 0.17 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr17_+_34423054 | 0.16 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr15_-_44291699 | 0.16 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr19_+_46345319 | 0.16 |
ENSMUST00000086969.13
|
Mfsd13a
|
major facilitator superfamily domain containing 13a |
| chr16_-_91723870 | 0.16 |
ENSMUST00000159295.8
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr5_-_137530214 | 0.16 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr17_+_32840283 | 0.16 |
ENSMUST00000077639.7
|
Cyp4f37
|
cytochrome P450, family 4, subfamily f, polypeptide 37 |
| chrX_+_135171002 | 0.15 |
ENSMUST00000178632.8
ENSMUST00000053540.11 |
Bex3
|
brain expressed X-linked 3 |
| chr18_-_67378886 | 0.15 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr17_+_29899420 | 0.15 |
ENSMUST00000130052.9
|
Cmtr1
|
cap methyltransferase 1 |
| chr7_-_79882313 | 0.15 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr3_-_115923098 | 0.15 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr8_-_111808488 | 0.15 |
ENSMUST00000188466.7
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr11_+_115044966 | 0.15 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr8_-_111808674 | 0.15 |
ENSMUST00000039597.14
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr17_-_25104910 | 0.15 |
ENSMUST00000234928.2
|
Nubp2
|
nucleotide binding protein 2 |
| chr7_+_100142977 | 0.15 |
ENSMUST00000129324.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr6_+_138118565 | 0.15 |
ENSMUST00000118091.8
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr6_+_135339543 | 0.15 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_3939924 | 0.14 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr7_-_30814652 | 0.14 |
ENSMUST00000168884.8
ENSMUST00000108102.9 |
Hpn
|
hepsin |
| chr17_-_25104874 | 0.14 |
ENSMUST00000234597.2
|
Nubp2
|
nucleotide binding protein 2 |
| chr4_+_43506966 | 0.14 |
ENSMUST00000030183.10
|
Car9
|
carbonic anhydrase 9 |
| chr17_+_32755525 | 0.14 |
ENSMUST00000169591.8
ENSMUST00000003416.15 |
Cyp4f16
|
cytochrome P450, family 4, subfamily f, polypeptide 16 |
| chr5_+_137756407 | 0.14 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr3_-_107838895 | 0.14 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr4_+_133311665 | 0.14 |
ENSMUST00000030661.14
ENSMUST00000105899.2 |
Gpn2
|
GPN-loop GTPase 2 |
| chr9_+_54493784 | 0.14 |
ENSMUST00000167866.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr13_+_4241149 | 0.14 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr1_-_120047868 | 0.14 |
ENSMUST00000112648.8
|
Dbi
|
diazepam binding inhibitor |
| chr2_+_31360219 | 0.14 |
ENSMUST00000102840.5
|
Ass1
|
argininosuccinate synthetase 1 |
| chr7_+_142086880 | 0.13 |
ENSMUST00000210803.2
|
Mrpl23
|
mitochondrial ribosomal protein L23 |
| chr2_-_105229653 | 0.13 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr13_-_6686686 | 0.13 |
ENSMUST00000136585.2
|
Pfkp
|
phosphofructokinase, platelet |
| chr2_-_181223787 | 0.13 |
ENSMUST00000057816.15
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chrX_+_135171049 | 0.13 |
ENSMUST00000113112.2
|
Bex3
|
brain expressed X-linked 3 |
| chr1_-_88205233 | 0.13 |
ENSMUST00000065420.12
ENSMUST00000054674.15 |
Hjurp
|
Holliday junction recognition protein |
| chr4_+_43957677 | 0.13 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr2_-_92876398 | 0.13 |
ENSMUST00000111272.3
ENSMUST00000178666.8 ENSMUST00000147339.3 |
Prdm11
|
PR domain containing 11 |
| chr3_-_107667499 | 0.13 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr10_+_82695013 | 0.13 |
ENSMUST00000020484.9
ENSMUST00000219962.3 |
Txnrd1
|
thioredoxin reductase 1 |
| chr4_+_99603877 | 0.13 |
ENSMUST00000097961.9
ENSMUST00000107004.9 ENSMUST00000139799.3 |
Alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr2_+_152907875 | 0.13 |
ENSMUST00000238488.2
ENSMUST00000129377.8 ENSMUST00000109800.2 |
Ccm2l
|
cerebral cavernous malformation 2-like |
| chr19_+_6135013 | 0.13 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr16_+_33712305 | 0.13 |
ENSMUST00000232262.2
|
Itgb5
|
integrin beta 5 |
| chr10_-_21036792 | 0.13 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr8_+_95703728 | 0.12 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_-_106076389 | 0.12 |
ENSMUST00000140761.9
|
Ppm1m
|
protein phosphatase 1M |
| chr9_-_53521585 | 0.12 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr10_+_118094593 | 0.12 |
ENSMUST00000219565.2
|
Gm32802
|
predicted gene, 32802 |
| chr7_-_79882501 | 0.12 |
ENSMUST00000065163.15
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr4_-_129436465 | 0.12 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr8_-_68270870 | 0.12 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr10_+_19497740 | 0.12 |
ENSMUST00000036564.8
|
Il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr12_+_87490666 | 0.12 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr1_+_127234441 | 0.12 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr18_-_35795233 | 0.12 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr10_-_75757393 | 0.12 |
ENSMUST00000121304.2
ENSMUST00000000925.10 |
Smarcb1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
| chr17_+_24689366 | 0.11 |
ENSMUST00000053024.8
|
Pgp
|
phosphoglycolate phosphatase |
| chr3_-_63872189 | 0.11 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr5_+_102916637 | 0.11 |
ENSMUST00000112852.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_-_149978560 | 0.11 |
ENSMUST00000094538.6
ENSMUST00000109931.8 ENSMUST00000089207.13 |
Zfp120
|
zinc finger protein 120 |
| chr7_+_27307173 | 0.11 |
ENSMUST00000136962.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr9_-_15212849 | 0.11 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_-_78875657 | 0.11 |
ENSMUST00000073001.5
|
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr9_+_69360902 | 0.11 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr3_+_88523440 | 0.11 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr8_+_121394961 | 0.11 |
ENSMUST00000034276.13
ENSMUST00000181586.8 |
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chrX_-_106446928 | 0.10 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr3_-_131065658 | 0.10 |
ENSMUST00000029610.9
|
Hadh
|
hydroxyacyl-Coenzyme A dehydrogenase |
| chr3_-_104725581 | 0.10 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr5_-_100867520 | 0.10 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr11_-_72106418 | 0.10 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr8_-_68270936 | 0.10 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_-_56151334 | 0.10 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr3_-_104725853 | 0.10 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr4_+_45965327 | 0.10 |
ENSMUST00000107777.9
|
Tdrd7
|
tudor domain containing 7 |
| chr12_-_79030250 | 0.10 |
ENSMUST00000070174.14
|
Tmem229b
|
transmembrane protein 229B |
| chr7_+_27307422 | 0.10 |
ENSMUST00000142365.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr9_+_119168714 | 0.10 |
ENSMUST00000176351.8
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr10_-_90959817 | 0.09 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr3_-_89152320 | 0.09 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr3_+_51323383 | 0.09 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr7_-_80053063 | 0.09 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_-_150293406 | 0.09 |
ENSMUST00000109916.8
|
Zfp442
|
zinc finger protein 442 |
| chr17_+_35188888 | 0.09 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr11_-_69811347 | 0.09 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_+_4186391 | 0.09 |
ENSMUST00000075221.3
|
Osm
|
oncostatin M |
| chr11_+_49327451 | 0.09 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr7_+_19144950 | 0.09 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr19_-_46917661 | 0.09 |
ENSMUST00000236727.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr6_+_28423539 | 0.09 |
ENSMUST00000020717.12
ENSMUST00000169841.2 |
Arf5
|
ADP-ribosylation factor 5 |
| chr2_-_164675357 | 0.09 |
ENSMUST00000042775.5
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr7_+_100971034 | 0.09 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr3_-_104725535 | 0.09 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr1_-_156501860 | 0.09 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr16_-_17394495 | 0.09 |
ENSMUST00000231645.2
ENSMUST00000232226.2 ENSMUST00000232336.2 ENSMUST00000232385.2 ENSMUST00000231615.2 ENSMUST00000231283.2 ENSMUST00000172164.10 |
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| chr2_+_13578738 | 0.09 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr11_-_117764258 | 0.09 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr17_+_29537798 | 0.09 |
ENSMUST00000114701.10
|
Pi16
|
peptidase inhibitor 16 |
| chr17_-_45903410 | 0.09 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr3_+_27371168 | 0.09 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr5_+_115061293 | 0.09 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr10_-_21036824 | 0.08 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr2_+_164611812 | 0.08 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr2_+_164328375 | 0.08 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_+_118286898 | 0.08 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chrX_+_149829131 | 0.08 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr16_+_90017634 | 0.08 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr6_-_34153955 | 0.08 |
ENSMUST00000019143.9
|
Slc35b4
|
solute carrier family 35, member B4 |
| chr2_-_32341408 | 0.08 |
ENSMUST00000028160.15
ENSMUST00000113310.9 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr2_-_32314017 | 0.08 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr3_-_90297187 | 0.08 |
ENSMUST00000029541.12
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr3_-_129548954 | 0.08 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr14_+_45567245 | 0.08 |
ENSMUST00000022380.9
|
Psmc6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr4_+_15265798 | 0.08 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr11_-_78875689 | 0.08 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr10_-_81127057 | 0.08 |
ENSMUST00000045744.7
|
Tjp3
|
tight junction protein 3 |
| chr12_+_55445560 | 0.08 |
ENSMUST00000021412.9
|
Psma6
|
proteasome subunit alpha 6 |
| chr9_-_107167046 | 0.07 |
ENSMUST00000035194.8
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr3_+_135531834 | 0.07 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr19_-_46950737 | 0.07 |
ENSMUST00000168536.9
ENSMUST00000236783.2 ENSMUST00000238106.2 |
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr9_+_50768224 | 0.07 |
ENSMUST00000174628.8
ENSMUST00000034560.14 ENSMUST00000114437.9 ENSMUST00000175645.8 ENSMUST00000176349.8 ENSMUST00000176798.8 ENSMUST00000175640.8 |
Ppp2r1b
|
protein phosphatase 2, regulatory subunit A, beta |
| chr18_-_35795175 | 0.07 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr2_+_28417800 | 0.07 |
ENSMUST00000238699.2
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr1_-_88205185 | 0.07 |
ENSMUST00000147393.2
|
Hjurp
|
Holliday junction recognition protein |
| chr11_-_78402931 | 0.07 |
ENSMUST00000052566.8
|
Tmem199
|
transmembrane protein 199 |
| chr15_+_74828272 | 0.07 |
ENSMUST00000188042.2
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr17_-_35081129 | 0.07 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr9_-_119169053 | 0.07 |
ENSMUST00000035092.7
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr2_+_13579092 | 0.07 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr4_-_129121676 | 0.07 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr11_-_82881929 | 0.07 |
ENSMUST00000138797.2
|
Slfn9
|
schlafen 9 |
| chr2_+_90677499 | 0.07 |
ENSMUST00000136872.8
ENSMUST00000150232.8 ENSMUST00000111467.4 |
Mtch2
|
mitochondrial carrier 2 |
| chr8_-_95602952 | 0.07 |
ENSMUST00000046461.9
|
Dok4
|
docking protein 4 |
| chrX_-_149595873 | 0.07 |
ENSMUST00000131241.2
ENSMUST00000147152.3 ENSMUST00000143843.8 |
Maged2
|
MAGE family member D2 |
| chr19_+_4644425 | 0.06 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr4_-_116982804 | 0.06 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr3_-_63872079 | 0.06 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr5_-_23881353 | 0.06 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr15_-_42540363 | 0.06 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr10_-_118076685 | 0.06 |
ENSMUST00000219147.2
|
Gm32717
|
predicted gene, 32717 |
| chr1_-_120048667 | 0.06 |
ENSMUST00000151708.3
|
Dbi
|
diazepam binding inhibitor |
| chr3_+_95836558 | 0.06 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr2_+_118998235 | 0.06 |
ENSMUST00000057454.4
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr9_-_71070506 | 0.06 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfic_Nfib
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 0.3 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.1 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.4 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) cytosol to ER transport(GO:0046967) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.2 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.2 | GO:0019371 | negative regulation of norepinephrine secretion(GO:0010700) cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.3 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.3 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.2 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.2 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.0 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |