Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nfkb2
Z-value: 1.24
Transcription factors associated with Nfkb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfkb2
|
ENSMUSG00000025225.16 | nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb2 | mm39_v1_chr19_+_46294119_46294235 | 0.85 | 6.8e-02 | Click! |
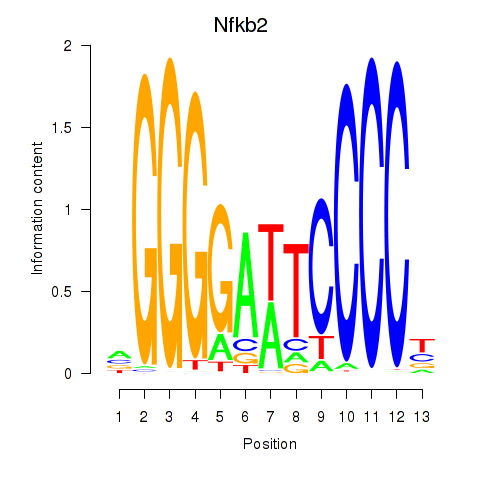
Activity profile of Nfkb2 motif
Sorted Z-values of Nfkb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90907207 | 1.02 |
ENSMUST00000031318.6
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr7_+_67925718 | 0.88 |
ENSMUST00000210558.2
|
Fam169b
|
family with sequence similarity 169, member B |
| chr4_+_118745806 | 0.64 |
ENSMUST00000106361.3
ENSMUST00000105035.3 |
Olfr1330
|
olfactory receptor 1330 |
| chr3_+_97565528 | 0.60 |
ENSMUST00000045743.13
|
Prkab2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr9_+_55234197 | 0.56 |
ENSMUST00000085754.10
ENSMUST00000034862.5 |
Tmem266
|
transmembrane protein 266 |
| chr1_-_5089564 | 0.54 |
ENSMUST00000002533.15
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr11_-_23720953 | 0.52 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr1_+_166828982 | 0.52 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr8_+_109441276 | 0.51 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr6_-_83433357 | 0.51 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr15_-_66158445 | 0.46 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chrX_+_162691978 | 0.45 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_+_77576981 | 0.40 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr1_+_166829001 | 0.38 |
ENSMUST00000126198.3
|
Fam78b
|
family with sequence similarity 78, member B |
| chr11_-_71928300 | 0.38 |
ENSMUST00000048207.10
|
Aipl1
|
aryl hydrocarbon receptor-interacting protein-like 1 |
| chr2_-_37312881 | 0.38 |
ENSMUST00000112936.4
ENSMUST00000112934.8 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr15_-_102154874 | 0.37 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma |
| chrX_+_162692126 | 0.37 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr7_-_44181477 | 0.37 |
ENSMUST00000098483.9
ENSMUST00000035323.6 |
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr7_+_3339059 | 0.35 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr8_-_106016097 | 0.33 |
ENSMUST00000171788.8
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr1_-_63153414 | 0.33 |
ENSMUST00000153992.2
ENSMUST00000165066.8 ENSMUST00000172416.8 ENSMUST00000137511.8 |
Ino80d
|
INO80 complex subunit D |
| chr4_-_125021610 | 0.30 |
ENSMUST00000036188.8
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr7_-_19363280 | 0.29 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr19_+_6391148 | 0.28 |
ENSMUST00000025897.13
ENSMUST00000130382.8 |
Map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr8_-_106015682 | 0.28 |
ENSMUST00000212922.2
ENSMUST00000212219.2 |
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr7_+_28988724 | 0.26 |
ENSMUST00000207714.2
ENSMUST00000048187.6 |
Ppp1r14a
|
protein phosphatase 1, regulatory inhibitor subunit 14A |
| chr2_-_5719302 | 0.26 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr17_+_35816915 | 0.22 |
ENSMUST00000025271.17
|
Pou5f1
|
POU domain, class 5, transcription factor 1 |
| chr1_-_66856695 | 0.21 |
ENSMUST00000068168.10
ENSMUST00000113987.2 |
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr1_-_152778373 | 0.21 |
ENSMUST00000073441.13
ENSMUST00000111836.4 |
Smg7
|
Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr10_+_80538482 | 0.20 |
ENSMUST00000095426.5
|
Izumo4
|
IZUMO family member 4 |
| chr7_+_30121147 | 0.20 |
ENSMUST00000108176.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr8_+_79754980 | 0.20 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr7_+_44866635 | 0.19 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr11_+_101066867 | 0.19 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr3_+_31149201 | 0.17 |
ENSMUST00000118470.8
ENSMUST00000029194.12 ENSMUST00000123532.2 |
Skil
|
SKI-like |
| chr8_-_105357941 | 0.16 |
ENSMUST00000034351.8
|
Rrad
|
Ras-related associated with diabetes |
| chr7_-_44702269 | 0.16 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chr7_-_100951227 | 0.16 |
ENSMUST00000122116.8
ENSMUST00000120267.9 |
Atg16l2
|
autophagy related 16-like 2 (S. cerevisiae) |
| chr3_+_31149247 | 0.15 |
ENSMUST00000117728.8
|
Skil
|
SKI-like |
| chr1_+_132996237 | 0.14 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr1_+_34840785 | 0.13 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chrX_-_7537580 | 0.12 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr1_-_36982747 | 0.12 |
ENSMUST00000185964.3
|
Tmem131
|
transmembrane protein 131 |
| chr10_+_127216459 | 0.11 |
ENSMUST00000166820.8
|
R3hdm2
|
R3H domain containing 2 |
| chr11_+_69920542 | 0.10 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_-_63153675 | 0.10 |
ENSMUST00000097718.9
|
Ino80d
|
INO80 complex subunit D |
| chr10_+_127216668 | 0.09 |
ENSMUST00000111628.9
|
R3hdm2
|
R3H domain containing 2 |
| chr17_-_36291087 | 0.08 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3 |
| chr11_-_54853729 | 0.07 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr10_-_89522112 | 0.07 |
ENSMUST00000092227.12
ENSMUST00000174252.8 |
Scyl2
|
SCY1-like 2 (S. cerevisiae) |
| chr7_+_30121776 | 0.07 |
ENSMUST00000108175.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr14_-_30348153 | 0.07 |
ENSMUST00000112211.9
ENSMUST00000112210.11 |
Prkcd
|
protein kinase C, delta |
| chr4_+_140815644 | 0.05 |
ENSMUST00000051907.3
|
Spata21
|
spermatogenesis associated 21 |
| chr8_+_124138163 | 0.04 |
ENSMUST00000071134.4
ENSMUST00000212743.2 |
Tubb3
|
tubulin, beta 3 class III |
| chr11_-_84761637 | 0.04 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr4_+_124880223 | 0.04 |
ENSMUST00000030687.8
|
Rspo1
|
R-spondin 1 |
| chr7_-_125799570 | 0.03 |
ENSMUST00000009344.16
|
Xpo6
|
exportin 6 |
| chr2_-_160155536 | 0.03 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr4_-_118615913 | 0.03 |
ENSMUST00000214922.3
ENSMUST00000216559.3 |
Olfr1338
|
olfactory receptor 1338 |
| chr7_-_43896112 | 0.03 |
ENSMUST00000237383.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr5_+_91039092 | 0.02 |
ENSMUST00000031327.9
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr2_-_164315634 | 0.02 |
ENSMUST00000017864.3
|
Trp53tg5
|
transformation related protein 53 target 5 |
| chr11_-_84761472 | 0.01 |
ENSMUST00000018547.9
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr11_+_70548622 | 0.01 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr6_+_68547717 | 0.00 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr8_-_71315651 | 0.00 |
ENSMUST00000212414.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfkb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.2 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.1 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.3 | GO:1903936 | miRNA catabolic process(GO:0010587) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.3 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.5 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.8 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |