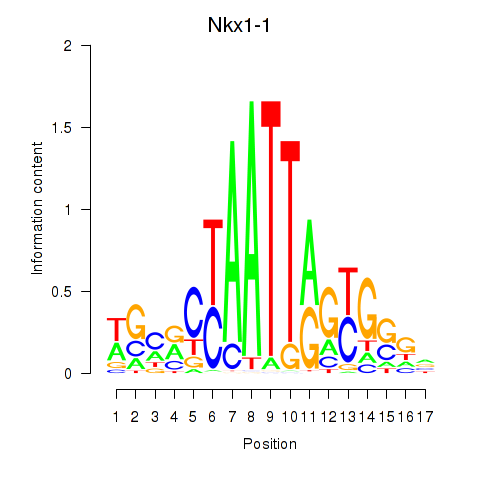
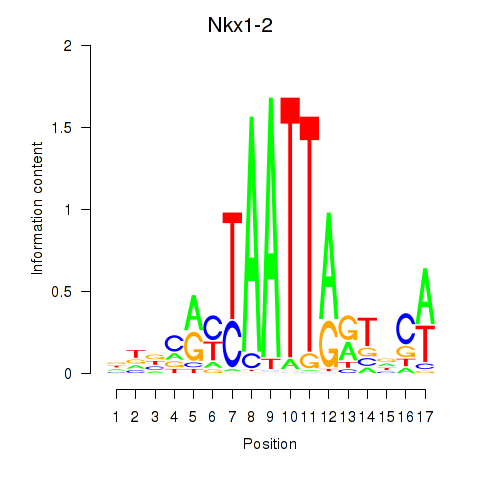
|
chr16_+_51851948
Show fit
|
0.94 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr16_+_51851917
Show fit
|
0.88 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr19_-_12313274
Show fit
|
0.75 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1
|
|
chr6_-_57340712
Show fit
|
0.71 |
ENSMUST00000228156.2
ENSMUST00000227186.2
ENSMUST00000228294.2
ENSMUST00000228342.2
|
Vmn1r17
|
vomeronasal 1 receptor 17
|
|
chr16_+_51851588
Show fit
|
0.61 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr2_-_89855921
Show fit
|
0.57 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264
|
|
chr5_-_84565218
Show fit
|
0.54 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5
|
|
chr8_-_3744167
Show fit
|
0.53 |
ENSMUST00000005678.6
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide
|
|
chr10_-_44024843
Show fit
|
0.49 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1
|
|
chr13_-_105191403
Show fit
|
0.48 |
ENSMUST00000063551.7
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein
|
|
chr17_-_37974666
Show fit
|
0.47 |
ENSMUST00000215414.2
ENSMUST00000213638.3
|
Olfr117
|
olfactory receptor 117
|
|
chr1_-_92412835
Show fit
|
0.46 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416
|
|
chr7_+_26895206
Show fit
|
0.44 |
ENSMUST00000179391.8
ENSMUST00000108379.8
|
BC024978
|
cDNA sequence BC024978
|
|
chr4_-_131802606
Show fit
|
0.42 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr6_+_57405915
Show fit
|
0.42 |
ENSMUST00000227909.2
|
Vmn1r20
|
vomeronasal 1 receptor 20
|
|
chr3_-_15902583
Show fit
|
0.41 |
ENSMUST00000108354.8
ENSMUST00000108349.2
ENSMUST00000108352.9
ENSMUST00000108350.8
ENSMUST00000050623.11
|
Sirpb1c
|
signal-regulatory protein beta 1C
|
|
chr2_-_111843053
Show fit
|
0.40 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310
|
|
chr10_-_128875765
Show fit
|
0.39 |
ENSMUST00000204763.3
|
Olfr764-ps1
|
olfactory receptor 764, pseudogene 1
|
|
chrX_+_159551009
Show fit
|
0.37 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human)
|
|
chr6_-_58195806
Show fit
|
0.37 |
ENSMUST00000228530.2
ENSMUST00000226666.2
|
Vmn1r27
|
vomeronasal 1 receptor 27
|
|
chr10_+_129539079
Show fit
|
0.36 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804
|
|
chr7_+_140181182
Show fit
|
0.35 |
ENSMUST00000214180.2
ENSMUST00000211771.2
|
Olfr46
|
olfactory receptor 46
|
|
chr2_-_89423470
Show fit
|
0.35 |
ENSMUST00000217254.2
ENSMUST00000217192.2
ENSMUST00000213221.2
|
Olfr1246
|
olfactory receptor 1246
|
|
chr1_+_173093568
Show fit
|
0.34 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418
|
|
chr7_+_3648264
Show fit
|
0.34 |
ENSMUST00000206287.2
ENSMUST00000038913.16
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr8_+_114362181
Show fit
|
0.33 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated
|
|
chrX_+_102400061
Show fit
|
0.33 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1
|
|
chr7_-_12829100
Show fit
|
0.32 |
ENSMUST00000209822.3
ENSMUST00000235753.2
|
Vmn1r85
|
vomeronasal 1 receptor 85
|
|
chr8_+_114362419
Show fit
|
0.29 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated
|
|
chr7_-_11414074
Show fit
|
0.29 |
ENSMUST00000227010.2
ENSMUST00000209638.3
|
Vmn1r72
|
vomeronasal 1 receptor 72
|
|
chr4_-_131802561
Show fit
|
0.27 |
ENSMUST00000105970.8
ENSMUST00000105975.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr1_-_91386976
Show fit
|
0.26 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2
|
|
chr7_-_100613579
Show fit
|
0.25 |
ENSMUST00000060174.6
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr7_-_73187369
Show fit
|
0.25 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2
|
|
chr11_+_121128042
Show fit
|
0.25 |
ENSMUST00000103015.4
|
Narf
|
nuclear prelamin A recognition factor
|
|
chrX_-_73129195
Show fit
|
0.22 |
ENSMUST00000140399.3
ENSMUST00000123362.8
ENSMUST00000100750.10
ENSMUST00000033770.13
|
Mecp2
|
methyl CpG binding protein 2
|
|
chr12_-_25146078
Show fit
|
0.22 |
ENSMUST00000222667.2
ENSMUST00000020974.7
|
Id2
|
inhibitor of DNA binding 2
|
|
chr19_+_12364643
Show fit
|
0.21 |
ENSMUST00000217062.3
ENSMUST00000216145.2
ENSMUST00000213657.2
|
Olfr1440
|
olfactory receptor 1440
|
|
chr11_-_99134885
Show fit
|
0.18 |
ENSMUST00000103132.10
ENSMUST00000038214.7
|
Krt222
|
keratin 222
|
|
chr1_-_150341911
Show fit
|
0.18 |
ENSMUST00000162367.8
ENSMUST00000161611.8
ENSMUST00000161320.8
ENSMUST00000159035.2
|
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein)
|
|
chr4_-_132191181
Show fit
|
0.18 |
ENSMUST00000102567.4
|
Med18
|
mediator complex subunit 18
|
|
chr2_-_160950936
Show fit
|
0.18 |
ENSMUST00000039782.14
ENSMUST00000134178.8
|
Chd6
|
chromodomain helicase DNA binding protein 6
|
|
chr9_-_107749594
Show fit
|
0.17 |
ENSMUST00000183035.2
|
Rbm6
|
RNA binding motif protein 6
|
|
chr14_-_52277310
Show fit
|
0.17 |
ENSMUST00000216907.2
ENSMUST00000214071.2
ENSMUST00000216188.2
|
Olfr221
|
olfactory receptor 221
|
|
chr4_-_43710231
Show fit
|
0.17 |
ENSMUST00000217544.2
ENSMUST00000107862.3
|
Olfr71
|
olfactory receptor 71
|
|
chr12_+_108145802
Show fit
|
0.17 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K
|
|
chr15_-_77840856
Show fit
|
0.16 |
ENSMUST00000117725.2
ENSMUST00000016696.13
|
Foxred2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr7_-_30251680
Show fit
|
0.16 |
ENSMUST00000215288.2
ENSMUST00000108165.8
ENSMUST00000153594.3
|
Proser3
|
proline and serine rich 3
|
|
chr6_+_29694181
Show fit
|
0.16 |
ENSMUST00000046750.14
ENSMUST00000115250.4
|
Tspan33
|
tetraspanin 33
|
|
chr8_+_84262409
Show fit
|
0.15 |
ENSMUST00000214156.2
ENSMUST00000209408.4
|
Olfr370
|
olfactory receptor 370
|
|
chr9_+_40092216
Show fit
|
0.15 |
ENSMUST00000218134.2
ENSMUST00000216720.2
ENSMUST00000214763.2
|
Olfr986
|
olfactory receptor 986
|
|
chr17_+_21446349
Show fit
|
0.15 |
ENSMUST00000235895.2
|
Vmn1r234
|
vomeronasal 1 receptor 234
|
|
chr4_-_141327146
Show fit
|
0.15 |
ENSMUST00000141518.8
ENSMUST00000127455.8
ENSMUST00000105784.8
|
Fblim1
|
filamin binding LIM protein 1
|
|
chr5_-_74692327
Show fit
|
0.15 |
ENSMUST00000072857.13
ENSMUST00000113542.9
ENSMUST00000151474.3
|
Scfd2
|
Sec1 family domain containing 2
|
|
chr19_+_11943265
Show fit
|
0.14 |
ENSMUST00000025590.11
|
Osbp
|
oxysterol binding protein
|
|
chr8_+_84699580
Show fit
|
0.14 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha
|
|
chr4_+_136038263
Show fit
|
0.12 |
ENSMUST00000105850.8
ENSMUST00000148843.10
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr6_-_128339458
Show fit
|
0.12 |
ENSMUST00000155573.3
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1
|
|
chr12_+_4819022
Show fit
|
0.12 |
ENSMUST00000219503.2
|
Pfn4
|
profilin family, member 4
|
|
chr5_-_143846600
Show fit
|
0.12 |
ENSMUST00000031613.11
ENSMUST00000100483.3
|
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2
|
|
chr18_+_4993795
Show fit
|
0.12 |
ENSMUST00000153016.8
|
Svil
|
supervillin
|
|
chrX_-_49108236
Show fit
|
0.12 |
ENSMUST00000213556.3
ENSMUST00000213463.2
|
Olfr1323
|
olfactory receptor 1323
|
|
chr11_-_97040858
Show fit
|
0.11 |
ENSMUST00000118375.8
|
Tbkbp1
|
TBK1 binding protein 1
|
|
chr19_+_5540483
Show fit
|
0.10 |
ENSMUST00000209469.2
ENSMUST00000116560.3
|
Cfl1
|
cofilin 1, non-muscle
|
|
chr6_-_58452315
Show fit
|
0.10 |
ENSMUST00000177318.3
ENSMUST00000176147.9
ENSMUST00000176023.3
ENSMUST00000228586.2
|
Vmn1r31
|
vomeronasal 1 receptor 31
|
|
chr14_-_70666513
Show fit
|
0.10 |
ENSMUST00000226426.2
ENSMUST00000048129.6
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2
|
|
chr4_+_136038301
Show fit
|
0.09 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr15_+_98350469
Show fit
|
0.09 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281
|
|
chr11_-_22236795
Show fit
|
0.09 |
ENSMUST00000180360.8
ENSMUST00000109563.9
|
Ehbp1
|
EH domain binding protein 1
|
|
chr4_+_136038243
Show fit
|
0.09 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr2_+_111355548
Show fit
|
0.08 |
ENSMUST00000217845.3
|
Olfr1293-ps
|
olfactory receptor 1293, pseudogene
|
|
chr19_+_29344846
Show fit
|
0.08 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen
|
|
chr17_-_57338468
Show fit
|
0.08 |
ENSMUST00000007814.10
ENSMUST00000233480.2
|
Khsrp
|
KH-type splicing regulatory protein
|
|
chr5_-_137101108
Show fit
|
0.07 |
ENSMUST00000077523.4
ENSMUST00000041388.11
|
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1
|
|
chr12_+_52746158
Show fit
|
0.07 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr2_-_45002902
Show fit
|
0.07 |
ENSMUST00000076836.13
ENSMUST00000176732.8
ENSMUST00000200844.4
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr9_-_26717686
Show fit
|
0.07 |
ENSMUST00000162702.8
ENSMUST00000040398.14
ENSMUST00000066560.13
|
Glb1l2
|
galactosidase, beta 1-like 2
|
|
chr12_-_4819216
Show fit
|
0.06 |
ENSMUST00000053458.7
ENSMUST00000218575.2
|
Fam228b
|
family with sequence similarity 228, member B
|
|
chr2_-_165230165
Show fit
|
0.06 |
ENSMUST00000103084.4
|
Zfp334
|
zinc finger protein 334
|
|
chr2_-_102230602
Show fit
|
0.06 |
ENSMUST00000152929.2
|
Trim44
|
tripartite motif-containing 44
|
|
chr7_-_26895141
Show fit
|
0.06 |
ENSMUST00000163311.9
ENSMUST00000126211.2
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr2_+_85804239
Show fit
|
0.06 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029
|
|
chr7_-_19363280
Show fit
|
0.06 |
ENSMUST00000094762.10
ENSMUST00000049912.15
ENSMUST00000098754.5
|
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B
|
|
chr16_+_13176238
Show fit
|
0.06 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B
|
|
chr5_+_143846782
Show fit
|
0.05 |
ENSMUST00000148011.8
ENSMUST00000110709.7
|
Pms2
|
PMS1 homolog2, mismatch repair system component
|
|
chr9_-_66033841
Show fit
|
0.05 |
ENSMUST00000137542.2
|
Snx1
|
sorting nexin 1
|
|
chr10_+_116137277
Show fit
|
0.05 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B
|
|
chr16_-_22676264
Show fit
|
0.05 |
ENSMUST00000232075.2
ENSMUST00000004576.8
|
Tbccd1
|
TBCC domain containing 1
|
|
chr4_-_88595161
Show fit
|
0.04 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16
|
|
chr4_-_141327253
Show fit
|
0.03 |
ENSMUST00000147785.8
|
Fblim1
|
filamin binding LIM protein 1
|
|
chrX_+_57076359
Show fit
|
0.03 |
ENSMUST00000088631.11
ENSMUST00000088629.4
|
Zic3
|
zinc finger protein of the cerebellum 3
|
|
chr1_+_173924457
Show fit
|
0.02 |
ENSMUST00000213832.2
|
Olfr427
|
olfactory receptor 427
|
|
chr6_+_37847721
Show fit
|
0.02 |
ENSMUST00000031859.14
ENSMUST00000120428.8
|
Trim24
|
tripartite motif-containing 24
|
|
chr6_+_145561483
Show fit
|
0.02 |
ENSMUST00000087445.7
|
Tuba3b
|
tubulin, alpha 3B
|
|
chr2_+_154390808
Show fit
|
0.02 |
ENSMUST00000045116.11
ENSMUST00000109709.4
|
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene
|
|
chr2_-_89678487
Show fit
|
0.02 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48
|
|
chr5_-_90788323
Show fit
|
0.02 |
ENSMUST00000202784.4
ENSMUST00000031317.10
ENSMUST00000201370.2
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chrX_+_57075981
Show fit
|
0.02 |
ENSMUST00000088627.11
|
Zic3
|
zinc finger protein of the cerebellum 3
|
|
chr5_+_9150691
Show fit
|
0.01 |
ENSMUST00000115365.3
|
Tmem243
|
transmembrane protein 243, mitochondrial
|
|
chr14_+_67148619
Show fit
|
0.01 |
ENSMUST00000089236.11
ENSMUST00000122431.3
|
Pnma2
|
paraneoplastic antigen MA2
|
|
chr16_+_22965286
Show fit
|
0.01 |
ENSMUST00000023593.6
|
Adipoq
|
adiponectin, C1Q and collagen domain containing
|
|
chr6_-_69704122
Show fit
|
0.01 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48
|
|
chr1_-_173161069
Show fit
|
0.01 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group)
|
|
chr6_-_128339775
Show fit
|
0.01 |
ENSMUST00000112152.8
ENSMUST00000057421.15
ENSMUST00000112151.2
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1
|
|
chr11_+_116734104
Show fit
|
0.01 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23
|
|
chr7_+_128346655
Show fit
|
0.01 |
ENSMUST00000042942.10
|
Sec23ip
|
Sec23 interacting protein
|
|
chr1_-_131441962
Show fit
|
0.01 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2
|
|
chr14_-_59602882
Show fit
|
0.00 |
ENSMUST00000160425.8
ENSMUST00000095157.11
|
Phf11d
|
PHD finger protein 11D
|
|
chr14_-_70666821
Show fit
|
0.00 |
ENSMUST00000226229.2
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2
|