Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
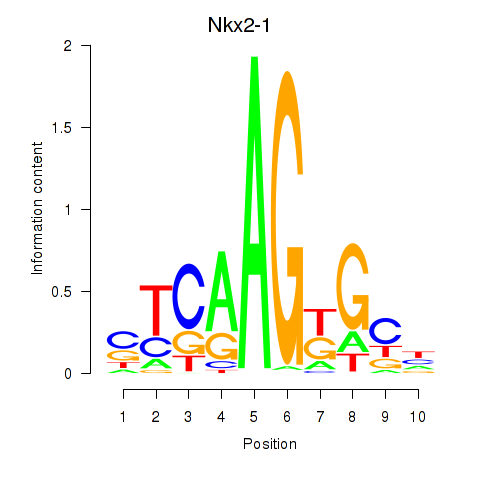
Results for Nkx2-1
Z-value: 1.16
Transcription factors associated with Nkx2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-1
|
ENSMUSG00000001496.16 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-1 | mm39_v1_chr12_-_56581823_56581891 | 0.83 | 8.5e-02 | Click! |
Activity profile of Nkx2-1 motif
Sorted Z-values of Nkx2-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23922783 | 1.19 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr17_-_57289121 | 1.00 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chr5_-_97259447 | 0.92 |
ENSMUST00000069453.9
ENSMUST00000112968.2 |
Paqr3
|
progestin and adipoQ receptor family member III |
| chr11_-_96868483 | 0.89 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr17_+_69071546 | 0.82 |
ENSMUST00000233625.2
|
L3mbtl4
|
L3MBTL4 histone methyl-lysine binding protein |
| chr10_-_67972401 | 0.77 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr7_+_28466160 | 0.72 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr2_+_71811526 | 0.71 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_128971620 | 0.66 |
ENSMUST00000035481.5
|
Chchd5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr11_+_32483290 | 0.65 |
ENSMUST00000102821.4
|
Stk10
|
serine/threonine kinase 10 |
| chr9_+_45924105 | 0.65 |
ENSMUST00000126865.8
|
Sik3
|
SIK family kinase 3 |
| chr3_+_90201388 | 0.63 |
ENSMUST00000199607.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr7_-_28465870 | 0.59 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr11_+_68989763 | 0.57 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr16_+_24266829 | 0.57 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr12_-_111638722 | 0.57 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr9_+_45924120 | 0.53 |
ENSMUST00000120463.9
ENSMUST00000120247.8 |
Sik3
|
SIK family kinase 3 |
| chr2_-_88937158 | 0.52 |
ENSMUST00000099806.3
ENSMUST00000213288.2 |
Olfr1220
|
olfactory receptor 1220 |
| chr13_+_38009981 | 0.50 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr17_-_34846323 | 0.47 |
ENSMUST00000168709.3
ENSMUST00000064953.15 ENSMUST00000170345.8 ENSMUST00000171121.9 ENSMUST00000168391.9 ENSMUST00000169067.9 |
Gm20460
Ppt2
|
predicted gene 20460 palmitoyl-protein thioesterase 2 |
| chrX_-_73009933 | 0.47 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr2_-_73722874 | 0.43 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chr9_+_75682637 | 0.42 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chr7_+_127845984 | 0.40 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr16_+_90535212 | 0.38 |
ENSMUST00000038197.3
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr2_-_73722932 | 0.36 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chrX_+_55391749 | 0.35 |
ENSMUST00000101560.4
|
Zfp449
|
zinc finger protein 449 |
| chr2_-_121948845 | 0.33 |
ENSMUST00000036450.8
|
Spg11
|
SPG11, spatacsin vesicle trafficking associated |
| chr1_-_173363523 | 0.32 |
ENSMUST00000139092.8
|
Ifi214
|
interferon activated gene 214 |
| chr11_+_70323452 | 0.32 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr7_-_30262659 | 0.31 |
ENSMUST00000043898.8
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr7_-_28824440 | 0.31 |
ENSMUST00000214374.2
ENSMUST00000179893.9 ENSMUST00000032813.10 |
Ryr1
|
ryanodine receptor 1, skeletal muscle |
| chr5_-_97259424 | 0.30 |
ENSMUST00000112969.10
|
Paqr3
|
progestin and adipoQ receptor family member III |
| chr11_-_118139312 | 0.29 |
ENSMUST00000100181.11
|
Cyth1
|
cytohesin 1 |
| chr12_+_85871404 | 0.28 |
ENSMUST00000177188.8
ENSMUST00000095536.10 ENSMUST00000110220.9 ENSMUST00000040179.14 |
Ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr1_+_74193138 | 0.28 |
ENSMUST00000027372.8
ENSMUST00000106899.4 |
Cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr7_-_4755971 | 0.28 |
ENSMUST00000183971.8
ENSMUST00000182173.2 ENSMUST00000182738.8 ENSMUST00000182111.8 ENSMUST00000184143.8 ENSMUST00000182048.2 ENSMUST00000063324.14 |
Cox6b2
|
cytochrome c oxidase subunit 6B2 |
| chr9_+_40092216 | 0.27 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr6_+_113303929 | 0.27 |
ENSMUST00000032406.15
|
Ogg1
|
8-oxoguanine DNA-glycosylase 1 |
| chr6_+_72281587 | 0.26 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr13_+_110039620 | 0.24 |
ENSMUST00000120664.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_-_83353787 | 0.24 |
ENSMUST00000021020.13
ENSMUST00000119346.2 ENSMUST00000103209.10 ENSMUST00000108137.9 |
Mmp28
|
matrix metallopeptidase 28 (epilysin) |
| chr1_+_178356678 | 0.21 |
ENSMUST00000161017.8
|
Kif26b
|
kinesin family member 26B |
| chr15_-_64254754 | 0.21 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr11_+_69920849 | 0.21 |
ENSMUST00000143920.4
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_+_153750081 | 0.20 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chrX_+_56008685 | 0.19 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr17_+_6157154 | 0.18 |
ENSMUST00000149756.8
|
Tulp4
|
tubby like protein 4 |
| chr6_-_38276157 | 0.18 |
ENSMUST00000058524.3
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like |
| chrX_+_72935708 | 0.18 |
ENSMUST00000033765.8
ENSMUST00000101470.3 ENSMUST00000114395.2 |
Avpr2
|
arginine vasopressin receptor 2 |
| chrX_+_7628891 | 0.18 |
ENSMUST00000077680.10
ENSMUST00000079542.13 ENSMUST00000115679.8 ENSMUST00000137467.8 |
Tfe3
|
transcription factor E3 |
| chr17_-_34847354 | 0.17 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_+_22520910 | 0.17 |
ENSMUST00000042261.5
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr11_+_101046708 | 0.16 |
ENSMUST00000043654.10
|
Tubg2
|
tubulin, gamma 2 |
| chr1_-_80191649 | 0.15 |
ENSMUST00000058748.2
|
Fam124b
|
family with sequence similarity 124, member B |
| chr11_+_80319424 | 0.14 |
ENSMUST00000173938.8
ENSMUST00000017572.14 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr19_-_8816530 | 0.14 |
ENSMUST00000096259.6
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr17_-_26204621 | 0.14 |
ENSMUST00000041641.9
ENSMUST00000212520.2 |
Capn15
|
calpain 15 |
| chr18_+_49965689 | 0.13 |
ENSMUST00000180611.8
|
Dmxl1
|
Dmx-like 1 |
| chr2_-_121304521 | 0.12 |
ENSMUST00000056732.4
|
Mfap1b
|
microfibrillar-associated protein 1B |
| chr7_-_30443106 | 0.12 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_+_68858942 | 0.12 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr11_+_69920956 | 0.11 |
ENSMUST00000232115.2
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr13_+_12580772 | 0.11 |
ENSMUST00000220811.2
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr12_+_91367764 | 0.11 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr11_-_118139331 | 0.11 |
ENSMUST00000017276.14
|
Cyth1
|
cytohesin 1 |
| chr2_-_89779008 | 0.11 |
ENSMUST00000214846.2
|
Olfr1259
|
olfactory receptor 1259 |
| chr17_+_48080113 | 0.10 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr11_+_74540284 | 0.09 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr2_-_57004933 | 0.09 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_+_43973167 | 0.09 |
ENSMUST00000087933.10
ENSMUST00000188313.7 |
Tpp2
|
tripeptidyl peptidase II |
| chr11_-_96807192 | 0.08 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr15_-_64254585 | 0.07 |
ENSMUST00000176384.8
ENSMUST00000175799.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr2_+_177760768 | 0.07 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr1_-_162726234 | 0.07 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr7_+_34885782 | 0.07 |
ENSMUST00000135452.8
ENSMUST00000001854.12 |
Slc7a10
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 10 |
| chrX_+_57075981 | 0.06 |
ENSMUST00000088627.11
|
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr13_+_38009951 | 0.06 |
ENSMUST00000138043.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr3_+_88439616 | 0.06 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr7_-_45463701 | 0.06 |
ENSMUST00000210898.2
ENSMUST00000107729.10 ENSMUST00000056820.13 |
Cyth2
|
cytohesin 2 |
| chr1_-_136273811 | 0.05 |
ENSMUST00000048309.12
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chrX_+_57076359 | 0.05 |
ENSMUST00000088631.11
ENSMUST00000088629.4 |
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr6_+_127554931 | 0.05 |
ENSMUST00000071563.5
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr4_+_62278932 | 0.05 |
ENSMUST00000084526.12
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr16_-_92622659 | 0.05 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr15_+_76579885 | 0.05 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr14_-_14651778 | 0.04 |
ENSMUST00000052678.9
|
Flnb
|
filamin, beta |
| chr18_-_68562385 | 0.04 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr13_+_55782814 | 0.04 |
ENSMUST00000172272.8
ENSMUST00000099479.4 ENSMUST00000223736.2 |
Ddx46
|
DEAD box helicase 46 |
| chr18_+_24842493 | 0.03 |
ENSMUST00000037097.9
ENSMUST00000234526.2 ENSMUST00000234834.2 |
Fhod3
|
formin homology 2 domain containing 3 |
| chr15_+_80118269 | 0.03 |
ENSMUST00000229138.2
ENSMUST00000166030.2 |
Mief1
|
mitochondrial elongation factor 1 |
| chr11_-_118139418 | 0.03 |
ENSMUST00000106305.9
|
Cyth1
|
cytohesin 1 |
| chr9_+_110075133 | 0.03 |
ENSMUST00000199736.2
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chrX_+_143471903 | 0.03 |
ENSMUST00000112843.2
|
Rtl4
|
retrotransposon Gag like 4 |
| chr2_-_73605684 | 0.02 |
ENSMUST00000112024.10
ENSMUST00000180045.8 |
Chn1
|
chimerin 1 |
| chr8_-_13446769 | 0.02 |
ENSMUST00000033826.4
|
Atp4b
|
ATPase, H+/K+ exchanging, beta polypeptide |
| chr14_+_67470884 | 0.02 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr13_+_105218625 | 0.02 |
ENSMUST00000022232.6
|
Nt5el
|
5' nucleotidase, ecto-like |
| chr19_-_5518515 | 0.02 |
ENSMUST00000236256.2
ENSMUST00000025844.6 |
Ctsw
|
cathepsin W |
| chrX_-_110372843 | 0.02 |
ENSMUST00000096348.10
ENSMUST00000113428.9 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr19_-_5452521 | 0.02 |
ENSMUST00000235569.2
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr14_+_26959975 | 0.02 |
ENSMUST00000049206.6
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr16_-_38649107 | 0.02 |
ENSMUST00000122078.3
|
Tex55
|
testis expressed 55 |
| chr5_+_92135294 | 0.02 |
ENSMUST00000178614.3
|
Odaph
|
odontogenesis associated phosphoprotein |
| chr14_-_24054352 | 0.01 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_+_96183294 | 0.01 |
ENSMUST00000173432.3
|
Hoxb6
|
homeobox B6 |
| chr17_+_6156738 | 0.01 |
ENSMUST00000142030.8
|
Tulp4
|
tubby like protein 4 |
| chr9_-_121621544 | 0.01 |
ENSMUST00000035110.11
|
Hhatl
|
hedgehog acyltransferase-like |
| chr15_-_7427815 | 0.01 |
ENSMUST00000096494.5
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chrX_-_110372800 | 0.01 |
ENSMUST00000137712.9
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr12_-_11200306 | 0.01 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr17_-_34847013 | 0.01 |
ENSMUST00000166040.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chrX_-_110372733 | 0.01 |
ENSMUST00000065976.12
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr18_+_37397991 | 0.01 |
ENSMUST00000052366.5
|
Pcdhb1
|
protocadherin beta 1 |
| chr15_-_7427759 | 0.01 |
ENSMUST00000058593.10
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr19_+_44481901 | 0.01 |
ENSMUST00000041163.5
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr5_-_145020840 | 0.01 |
ENSMUST00000151196.2
|
Kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr11_-_120344299 | 0.01 |
ENSMUST00000026452.3
|
Pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr1_+_85945790 | 0.01 |
ENSMUST00000149469.3
|
Spata3
|
spermatogenesis associated 3 |
| chr9_-_4796217 | 0.01 |
ENSMUST00000027020.13
ENSMUST00000163309.2 |
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr18_-_66993567 | 0.01 |
ENSMUST00000057942.4
|
Mc4r
|
melanocortin 4 receptor |
| chr7_-_34974953 | 0.01 |
ENSMUST00000238407.2
|
Wdr88
|
WD repeat domain 88 |
| chr4_-_44704006 | 0.01 |
ENSMUST00000146335.8
|
Pax5
|
paired box 5 |
| chr1_-_155617773 | 0.00 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr18_+_37276556 | 0.00 |
ENSMUST00000047479.3
|
Pcdhac2
|
protocadherin alpha subfamily C, 2 |
| chr2_-_26098293 | 0.00 |
ENSMUST00000054099.16
|
Lhx3
|
LIM homeobox protein 3 |
| chr1_+_75427080 | 0.00 |
ENSMUST00000113577.8
|
Asic4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr11_+_69920542 | 0.00 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_+_75427343 | 0.00 |
ENSMUST00000037708.10
|
Asic4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr9_-_4796142 | 0.00 |
ENSMUST00000063508.15
|
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr4_-_44703413 | 0.00 |
ENSMUST00000186542.2
|
Pax5
|
paired box 5 |
| chr2_-_168554774 | 0.00 |
ENSMUST00000123156.8
ENSMUST00000156555.2 |
Atp9a
|
ATPase, class II, type 9A |
| chr10_-_62723238 | 0.00 |
ENSMUST00000228901.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr9_+_110595224 | 0.00 |
ENSMUST00000136695.3
|
Myl3
|
myosin, light polypeptide 3 |
| chr9_-_37058590 | 0.00 |
ENSMUST00000080754.12
ENSMUST00000188057.7 ENSMUST00000039674.13 |
Pknox2
|
Pbx/knotted 1 homeobox 2 |
| chr6_+_113449237 | 0.00 |
ENSMUST00000204447.3
|
Il17rc
|
interleukin 17 receptor C |
| chr6_-_40864428 | 0.00 |
ENSMUST00000031937.4
|
Moxd2
|
monooxygenase, DBH-like 2 |
| chr7_-_144761806 | 0.00 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr2_-_170248421 | 0.00 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr9_-_62029936 | 0.00 |
ENSMUST00000185873.2
|
Glce
|
glucuronyl C5-epimerase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 1.0 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.2 | 0.8 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.7 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.4 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 0.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.8 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 1.2 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.3 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 1.1 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.0 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0048188 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.4 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |