Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
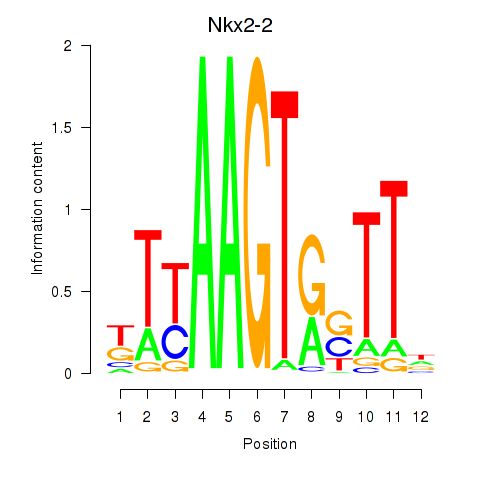
Results for Nkx2-2
Z-value: 0.22
Transcription factors associated with Nkx2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-2
|
ENSMUSG00000027434.12 | NK2 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-2 | mm39_v1_chr2_-_147028309_147028331 | -0.77 | 1.3e-01 | Click! |
Activity profile of Nkx2-2 motif
Sorted Z-values of Nkx2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_113237505 | 0.13 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr15_-_74869684 | 0.13 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr15_+_98065039 | 0.13 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr7_-_43956326 | 0.12 |
ENSMUST00000004587.11
|
Clec11a
|
C-type lectin domain family 11, member a |
| chr10_+_87926932 | 0.11 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr11_+_23234644 | 0.11 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr3_-_130524024 | 0.11 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr3_-_57559088 | 0.10 |
ENSMUST00000160959.8
|
Commd2
|
COMM domain containing 2 |
| chr5_-_110987441 | 0.10 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr10_-_117681864 | 0.09 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr11_+_52265090 | 0.09 |
ENSMUST00000020673.3
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr5_-_145128321 | 0.09 |
ENSMUST00000161845.2
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
| chr2_+_140012560 | 0.09 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr6_+_137731526 | 0.08 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr7_+_101060093 | 0.08 |
ENSMUST00000084894.15
|
Gm45837
|
predicted gene 45837 |
| chr11_+_48691175 | 0.08 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr14_-_56026266 | 0.08 |
ENSMUST00000168716.8
ENSMUST00000178399.3 ENSMUST00000022830.14 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr1_+_79753735 | 0.07 |
ENSMUST00000027464.9
|
Mrpl44
|
mitochondrial ribosomal protein L44 |
| chr7_-_132178101 | 0.07 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr6_+_88442391 | 0.07 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr12_+_76353835 | 0.07 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr2_+_170353338 | 0.07 |
ENSMUST00000136839.2
ENSMUST00000109148.8 ENSMUST00000170167.8 |
Pfdn4
|
prefoldin 4 |
| chrX_+_74425990 | 0.07 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr6_+_40619913 | 0.07 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr10_+_22236451 | 0.07 |
ENSMUST00000182677.8
|
Raet1d
|
retinoic acid early transcript delta |
| chr14_-_101846459 | 0.07 |
ENSMUST00000161991.8
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr12_+_105996961 | 0.07 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr2_-_150450402 | 0.07 |
ENSMUST00000046399.7
|
Apmap
|
adipocyte plasma membrane associated protein |
| chrM_+_8603 | 0.07 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_-_34847354 | 0.07 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr3_+_94350622 | 0.07 |
ENSMUST00000029786.14
ENSMUST00000196143.2 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr11_-_90281721 | 0.07 |
ENSMUST00000004051.8
|
Hlf
|
hepatic leukemia factor |
| chr10_-_93727003 | 0.07 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr12_+_33003882 | 0.07 |
ENSMUST00000076698.13
|
Sypl
|
synaptophysin-like protein |
| chr7_+_46490899 | 0.07 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr3_-_146475974 | 0.06 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr18_-_36859732 | 0.06 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr3_-_146476331 | 0.06 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr10_-_88601443 | 0.06 |
ENSMUST00000220188.2
ENSMUST00000218967.2 |
Utp20
|
UTP20 small subunit processome component |
| chr4_+_102843540 | 0.06 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr6_+_137731599 | 0.06 |
ENSMUST00000204356.2
|
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr19_-_60770628 | 0.06 |
ENSMUST00000238125.2
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr17_+_18108102 | 0.06 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr3_-_107893676 | 0.06 |
ENSMUST00000066530.7
ENSMUST00000012348.9 |
Gstm2
|
glutathione S-transferase, mu 2 |
| chr6_+_85428464 | 0.06 |
ENSMUST00000032078.9
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr18_-_61919707 | 0.06 |
ENSMUST00000120472.2
|
Afap1l1
|
actin filament associated protein 1-like 1 |
| chr11_-_21320452 | 0.06 |
ENSMUST00000102875.11
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr17_+_28910302 | 0.06 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chrX_+_106192510 | 0.06 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr9_-_106768601 | 0.06 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr12_-_79211820 | 0.06 |
ENSMUST00000162569.8
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr16_-_30152708 | 0.06 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr10_-_75609185 | 0.06 |
ENSMUST00000001716.8
|
Ddt
|
D-dopachrome tautomerase |
| chr8_+_40807344 | 0.06 |
ENSMUST00000136835.2
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr9_+_104930438 | 0.05 |
ENSMUST00000149243.8
ENSMUST00000035177.15 ENSMUST00000214036.2 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr7_+_19079058 | 0.05 |
ENSMUST00000160369.8
|
Ercc1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 |
| chr2_-_127286444 | 0.05 |
ENSMUST00000028848.4
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr7_-_26866157 | 0.05 |
ENSMUST00000080058.11
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr6_+_94477294 | 0.05 |
ENSMUST00000061118.11
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr2_+_130509530 | 0.05 |
ENSMUST00000103193.5
|
Itpa
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr2_-_152857239 | 0.05 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr17_-_34847013 | 0.05 |
ENSMUST00000166040.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_+_78192355 | 0.05 |
ENSMUST00000045026.4
|
Spag5
|
sperm associated antigen 5 |
| chr6_-_108162513 | 0.05 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr3_-_104771716 | 0.05 |
ENSMUST00000195912.2
ENSMUST00000094028.10 |
Capza1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chr5_-_124492734 | 0.05 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr1_-_80255156 | 0.05 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr14_+_76714350 | 0.05 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr6_+_67873135 | 0.05 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr3_-_94949854 | 0.05 |
ENSMUST00000117355.2
ENSMUST00000071664.12 ENSMUST00000107237.8 |
Psmd4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr12_+_73948143 | 0.04 |
ENSMUST00000110461.8
|
Hif1a
|
hypoxia inducible factor 1, alpha subunit |
| chr2_+_30697838 | 0.04 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr10_-_53952686 | 0.04 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr17_+_79359617 | 0.04 |
ENSMUST00000233916.2
|
Qpct
|
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
| chrX_+_141608694 | 0.04 |
ENSMUST00000112888.2
|
Tmem164
|
transmembrane protein 164 |
| chr2_+_173579285 | 0.04 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr11_-_73215442 | 0.04 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr13_-_20008397 | 0.04 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr13_+_34148691 | 0.04 |
ENSMUST00000021843.13
|
Nqo2
|
N-ribosyldihydronicotinamide quinone reductase 2 |
| chr1_-_69723316 | 0.04 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr1_-_152262339 | 0.04 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr15_-_10470575 | 0.04 |
ENSMUST00000136591.8
|
Dnajc21
|
DnaJ heat shock protein family (Hsp40) member C21 |
| chr2_+_83474779 | 0.04 |
ENSMUST00000081591.7
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr17_-_15784582 | 0.04 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr5_-_110987604 | 0.04 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr11_-_70895213 | 0.04 |
ENSMUST00000124464.2
ENSMUST00000108527.8 |
Dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr7_+_120634834 | 0.04 |
ENSMUST00000207351.2
|
Mettl9
|
methyltransferase like 9 |
| chr2_+_43638814 | 0.04 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr18_-_84969601 | 0.04 |
ENSMUST00000025547.4
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr8_+_46984016 | 0.04 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr16_+_23338960 | 0.04 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr13_+_104315301 | 0.04 |
ENSMUST00000022225.12
ENSMUST00000069187.12 |
Trim23
|
tripartite motif-containing 23 |
| chrX_+_67864699 | 0.03 |
ENSMUST00000096420.3
|
Gm14698
|
predicted gene 14698 |
| chr9_+_108225026 | 0.03 |
ENSMUST00000035237.12
ENSMUST00000194959.6 |
Usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr18_+_6765145 | 0.03 |
ENSMUST00000234810.2
ENSMUST00000234626.2 ENSMUST00000097680.7 |
Rab18
|
RAB18, member RAS oncogene family |
| chr2_+_173918089 | 0.03 |
ENSMUST00000155000.8
ENSMUST00000134876.8 ENSMUST00000147038.8 |
Stx16
|
syntaxin 16 |
| chr14_+_54923655 | 0.03 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr5_-_145128376 | 0.03 |
ENSMUST00000037056.9
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
| chr5_-_69749617 | 0.03 |
ENSMUST00000173927.8
ENSMUST00000120789.8 ENSMUST00000031117.13 |
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr16_-_57427179 | 0.03 |
ENSMUST00000114371.5
ENSMUST00000232413.2 |
Cmss1
|
cms small ribosomal subunit 1 |
| chr17_-_23990479 | 0.03 |
ENSMUST00000086325.13
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr15_-_36792649 | 0.03 |
ENSMUST00000126184.2
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr11_-_79971750 | 0.03 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr2_+_21210527 | 0.03 |
ENSMUST00000054591.10
ENSMUST00000102952.8 ENSMUST00000138965.8 ENSMUST00000138914.8 ENSMUST00000102951.2 |
Thnsl1
|
threonine synthase-like 1 (bacterial) |
| chr8_-_23747023 | 0.03 |
ENSMUST00000121783.7
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr6_-_120334400 | 0.03 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr5_-_3646071 | 0.03 |
ENSMUST00000121877.3
|
Rbm48
|
RNA binding motif protein 48 |
| chr1_-_66902429 | 0.03 |
ENSMUST00000027153.6
|
Acadl
|
acyl-Coenzyme A dehydrogenase, long-chain |
| chr19_-_46950948 | 0.03 |
ENSMUST00000236924.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr2_-_26127360 | 0.03 |
ENSMUST00000036187.9
|
Qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chrX_-_100463810 | 0.03 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr1_+_179788675 | 0.03 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr8_+_107757847 | 0.03 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr11_-_69791756 | 0.03 |
ENSMUST00000018714.13
ENSMUST00000128046.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr19_+_44551280 | 0.03 |
ENSMUST00000040455.5
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chrX_-_100463395 | 0.03 |
ENSMUST00000117901.8
ENSMUST00000120201.8 ENSMUST00000117637.8 ENSMUST00000134005.2 ENSMUST00000121520.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chrX_+_163763588 | 0.03 |
ENSMUST00000167446.8
ENSMUST00000057150.8 |
Fancb
|
Fanconi anemia, complementation group B |
| chr2_+_163503415 | 0.03 |
ENSMUST00000135537.8
|
Pkig
|
protein kinase inhibitor, gamma |
| chr19_-_6065872 | 0.03 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr6_-_70769135 | 0.03 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr16_+_4559426 | 0.03 |
ENSMUST00000120232.8
|
Hmox2
|
heme oxygenase 2 |
| chr13_+_14804739 | 0.03 |
ENSMUST00000178289.3
ENSMUST00000038690.6 ENSMUST00000222052.2 |
AW209491
|
expressed sequence AW209491 |
| chr10_-_107747995 | 0.03 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr11_-_115078147 | 0.03 |
ENSMUST00000103038.8
ENSMUST00000103039.2 ENSMUST00000103040.11 |
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr15_-_102259158 | 0.02 |
ENSMUST00000231061.2
ENSMUST00000041208.9 |
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr10_-_89568106 | 0.02 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr2_-_39116044 | 0.02 |
ENSMUST00000204368.2
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr1_-_152262425 | 0.02 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr3_+_137849608 | 0.02 |
ENSMUST00000159481.8
ENSMUST00000161141.2 |
Trmt10a
|
tRNA methyltransferase 10A |
| chr7_+_23874457 | 0.02 |
ENSMUST00000205309.2
ENSMUST00000206547.2 |
Zfp114
|
zinc finger protein 114 |
| chrX_+_106193167 | 0.02 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr2_+_22959223 | 0.02 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr10_-_59277570 | 0.02 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr14_-_63415235 | 0.02 |
ENSMUST00000054963.10
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr9_+_3335135 | 0.02 |
ENSMUST00000212294.2
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr13_+_21663077 | 0.02 |
ENSMUST00000062609.6
ENSMUST00000225845.2 |
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr12_-_91815855 | 0.02 |
ENSMUST00000167466.2
ENSMUST00000021347.12 ENSMUST00000178462.8 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr11_+_96209093 | 0.02 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr13_+_34148918 | 0.02 |
ENSMUST00000223479.2
|
Nqo2
|
N-ribosyldihydronicotinamide quinone reductase 2 |
| chr17_+_25115461 | 0.02 |
ENSMUST00000024978.7
|
Nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr9_+_35470752 | 0.02 |
ENSMUST00000034615.10
ENSMUST00000121246.2 |
Pus3
|
pseudouridine synthase 3 |
| chr2_-_85992176 | 0.02 |
ENSMUST00000099908.6
ENSMUST00000215624.2 |
Olfr1042
|
olfactory receptor 1042 |
| chr5_+_102872838 | 0.02 |
ENSMUST00000112853.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_-_34326847 | 0.02 |
ENSMUST00000202042.2
ENSMUST00000060049.8 |
Haus3
|
HAUS augmin-like complex, subunit 3 |
| chr2_-_140012447 | 0.02 |
ENSMUST00000046030.8
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr8_-_26330468 | 0.02 |
ENSMUST00000110609.8
|
Ash2l
|
ASH2 like histone lysine methyltransferase complex subunit |
| chr3_+_40585435 | 0.02 |
ENSMUST00000061590.6
|
Intu
|
inturned planar cell polarity protein |
| chr8_+_22996233 | 0.02 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr11_-_69791774 | 0.02 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr16_-_33787399 | 0.02 |
ENSMUST00000023510.7
|
Umps
|
uridine monophosphate synthetase |
| chr11_-_97774945 | 0.02 |
ENSMUST00000093939.4
|
Fbxo47
|
F-box protein 47 |
| chr4_+_3940747 | 0.02 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_+_108185331 | 0.02 |
ENSMUST00000131656.2
|
Coa7
|
cytochrome c oxidase assembly factor 7 |
| chr7_+_127836502 | 0.02 |
ENSMUST00000044660.6
|
Armc5
|
armadillo repeat containing 5 |
| chr12_+_88050467 | 0.02 |
ENSMUST00000222340.2
|
Eif1ad18
|
eukaryotic translation initiation factor 1A domain containing 18 |
| chr13_-_67080968 | 0.02 |
ENSMUST00000172597.8
ENSMUST00000173773.2 |
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr2_+_173918715 | 0.02 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr11_-_57409423 | 0.02 |
ENSMUST00000108850.2
ENSMUST00000020831.13 |
Fam114a2
|
family with sequence similarity 114, member A2 |
| chr11_+_75521800 | 0.02 |
ENSMUST00000006286.9
|
Inpp5k
|
inositol polyphosphate 5-phosphatase K |
| chr11_-_120348762 | 0.02 |
ENSMUST00000137632.2
ENSMUST00000044007.3 |
Oxld1
|
oxidoreductase like domain containing 1 |
| chr7_-_103094646 | 0.02 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr1_-_97904958 | 0.02 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_174292471 | 0.02 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr9_-_119151428 | 0.02 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr5_-_90371816 | 0.02 |
ENSMUST00000118816.6
ENSMUST00000048363.10 |
Cox18
|
cytochrome c oxidase assembly protein 18 |
| chr14_-_104760051 | 0.02 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr11_+_88864753 | 0.02 |
ENSMUST00000036649.8
|
Coil
|
coilin |
| chr5_+_76677150 | 0.02 |
ENSMUST00000087133.11
ENSMUST00000113493.8 ENSMUST00000049469.13 |
Exoc1
|
exocyst complex component 1 |
| chr3_+_81839908 | 0.01 |
ENSMUST00000029649.3
|
Ctso
|
cathepsin O |
| chr18_+_37858753 | 0.01 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr19_-_32080496 | 0.01 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr15_+_21111428 | 0.01 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr6_-_120334382 | 0.01 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr19_-_11243530 | 0.01 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr9_+_105272507 | 0.01 |
ENSMUST00000035181.10
ENSMUST00000123807.8 |
Aste1
|
asteroid homolog 1 |
| chr11_-_115078653 | 0.01 |
ENSMUST00000103041.8
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_-_69791712 | 0.01 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr14_+_53994813 | 0.01 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr19_+_34078333 | 0.01 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr18_+_37807357 | 0.01 |
ENSMUST00000073447.8
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr5_-_8417982 | 0.01 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr13_-_76166789 | 0.01 |
ENSMUST00000179078.9
ENSMUST00000167271.9 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr13_+_24798591 | 0.01 |
ENSMUST00000091694.10
|
Ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr7_+_19741948 | 0.01 |
ENSMUST00000073151.13
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr6_-_131637112 | 0.01 |
ENSMUST00000065781.5
|
Tas2r107
|
taste receptor, type 2, member 107 |
| chr7_-_100504610 | 0.01 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr6_-_115815373 | 0.01 |
ENSMUST00000032468.12
ENSMUST00000184428.6 |
Efcab12
|
EF-hand calcium binding domain 12 |
| chr11_-_45845992 | 0.01 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr1_-_39616369 | 0.01 |
ENSMUST00000195705.2
|
Rnf149
|
ring finger protein 149 |
| chrX_+_163052367 | 0.01 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr6_+_14901343 | 0.01 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr9_-_83316656 | 0.01 |
ENSMUST00000186802.2
|
Lca5
|
Leber congenital amaurosis 5 (human) |
| chr3_+_5283577 | 0.01 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr6_-_120334302 | 0.01 |
ENSMUST00000163827.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr6_+_122967309 | 0.01 |
ENSMUST00000079379.3
|
Clec4a4
|
C-type lectin domain family 4, member a4 |
| chr3_-_49711765 | 0.01 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr4_+_124923785 | 0.01 |
ENSMUST00000030684.8
|
Gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr9_-_96601574 | 0.01 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chrX_+_106193060 | 0.01 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr17_+_43879496 | 0.01 |
ENSMUST00000169694.2
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0009257 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.0 | 0.1 | GO:0006296 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0046725 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.0 | 0.1 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.0 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |