Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
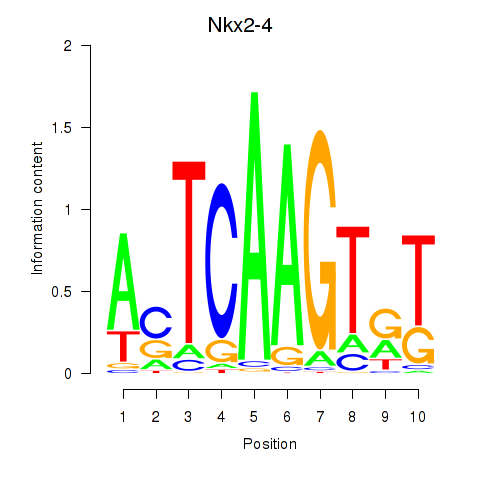
Results for Nkx2-4
Z-value: 0.66
Transcription factors associated with Nkx2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-4
|
ENSMUSG00000054160.3 | NK2 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-4 | mm39_v1_chr2_-_146927365_146927406 | 0.02 | 9.8e-01 | Click! |
Activity profile of Nkx2-4 motif
Sorted Z-values of Nkx2-4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_136620376 | 0.47 |
ENSMUST00000046332.6
|
C1qc
|
complement component 1, q subcomponent, C chain |
| chr13_+_63387827 | 0.45 |
ENSMUST00000222929.2
|
Aopep
|
aminopeptidase O |
| chr2_-_157408239 | 0.36 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr5_-_120887864 | 0.35 |
ENSMUST00000053909.13
ENSMUST00000081491.13 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr4_-_40279382 | 0.31 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chrX_+_55493325 | 0.31 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr9_+_108437485 | 0.29 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr1_+_16175998 | 0.27 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr18_+_36431732 | 0.27 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr13_+_63387870 | 0.26 |
ENSMUST00000159152.3
ENSMUST00000221820.2 |
Aopep
|
aminopeptidase O |
| chr8_-_79539838 | 0.26 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr10_+_4561974 | 0.26 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr2_+_140012560 | 0.24 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr11_+_49138278 | 0.24 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr2_-_34645241 | 0.24 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr8_+_78276020 | 0.22 |
ENSMUST00000056237.15
ENSMUST00000118622.2 |
Prmt9
|
protein arginine methyltransferase 9 |
| chr7_-_84328553 | 0.22 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr10_+_128067964 | 0.20 |
ENSMUST00000125289.8
ENSMUST00000105242.8 |
Timeless
|
timeless circadian clock 1 |
| chr5_-_65090104 | 0.19 |
ENSMUST00000059349.6
|
Tlr1
|
toll-like receptor 1 |
| chr12_+_33004178 | 0.18 |
ENSMUST00000020885.13
|
Sypl
|
synaptophysin-like protein |
| chr10_+_128067934 | 0.18 |
ENSMUST00000055539.11
ENSMUST00000105244.8 ENSMUST00000105243.9 |
Timeless
|
timeless circadian clock 1 |
| chr9_+_4376556 | 0.18 |
ENSMUST00000212075.2
|
Msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr5_-_137531463 | 0.17 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chrX_-_139857424 | 0.16 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr3_-_108469740 | 0.16 |
ENSMUST00000090546.6
|
Tmem167b
|
transmembrane protein 167B |
| chr1_+_85454323 | 0.15 |
ENSMUST00000239236.2
|
Gm7592
|
predicted gene 7592 |
| chrX_+_106132840 | 0.15 |
ENSMUST00000118666.8
ENSMUST00000053375.4 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr2_-_112089627 | 0.14 |
ENSMUST00000043970.2
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr5_-_137531413 | 0.14 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr15_+_88703786 | 0.13 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr14_+_48683581 | 0.12 |
ENSMUST00000227440.2
ENSMUST00000124720.8 ENSMUST00000226422.2 ENSMUST00000226400.2 |
Tmem260
|
transmembrane protein 260 |
| chr2_+_29951859 | 0.12 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chr11_-_103588487 | 0.11 |
ENSMUST00000107013.3
|
Gosr2
|
golgi SNAP receptor complex member 2 |
| chr4_-_129467430 | 0.11 |
ENSMUST00000102596.8
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr5_+_35156389 | 0.11 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr12_-_81468705 | 0.10 |
ENSMUST00000085319.4
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chr6_+_123206802 | 0.10 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr3_-_108469468 | 0.10 |
ENSMUST00000106622.3
|
Tmem167b
|
transmembrane protein 167B |
| chr14_+_55909816 | 0.10 |
ENSMUST00000227178.2
ENSMUST00000227914.2 |
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr6_-_29164981 | 0.10 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr12_+_105996961 | 0.09 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr15_+_99568208 | 0.09 |
ENSMUST00000023758.9
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr8_-_110305672 | 0.09 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr1_+_55170411 | 0.08 |
ENSMUST00000027122.14
|
Mob4
|
MOB family member 4, phocein |
| chr2_-_63014622 | 0.08 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr10_+_87926932 | 0.08 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr4_-_133599616 | 0.07 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr5_-_23821523 | 0.07 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr1_+_55170390 | 0.07 |
ENSMUST00000159311.8
ENSMUST00000162364.8 |
Mob4
|
MOB family member 4, phocein |
| chr1_+_59669482 | 0.07 |
ENSMUST00000190490.2
|
Gm973
|
predicted gene 973 |
| chr5_+_32293145 | 0.07 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr13_+_83723255 | 0.07 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr17_-_57501170 | 0.07 |
ENSMUST00000005976.8
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr19_-_6947076 | 0.06 |
ENSMUST00000237074.2
|
Plcb3
|
phospholipase C, beta 3 |
| chr1_+_121358778 | 0.06 |
ENSMUST00000036025.16
ENSMUST00000112621.2 |
Ccdc93
|
coiled-coil domain containing 93 |
| chr13_+_35843816 | 0.06 |
ENSMUST00000075220.14
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr1_-_138547403 | 0.06 |
ENSMUST00000027642.5
ENSMUST00000186017.7 |
Nek7
|
NIMA (never in mitosis gene a)-related expressed kinase 7 |
| chr11_+_87483723 | 0.06 |
ENSMUST00000119628.8
|
Mtmr4
|
myotubularin related protein 4 |
| chr3_+_36205166 | 0.06 |
ENSMUST00000197653.5
ENSMUST00000205077.2 |
Zfp267
|
zinc finger protein 267 |
| chr3_-_129548954 | 0.05 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr3_-_108108113 | 0.05 |
ENSMUST00000106655.2
ENSMUST00000065664.7 |
Cyb561d1
|
cytochrome b-561 domain containing 1 |
| chr5_+_35156454 | 0.05 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr12_+_86999366 | 0.05 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr19_-_6947112 | 0.05 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chr8_+_46944000 | 0.05 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr9_-_14815228 | 0.04 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr9_+_38738911 | 0.03 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr14_+_55909692 | 0.03 |
ENSMUST00000002397.7
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr15_+_21111428 | 0.03 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr3_+_88536805 | 0.03 |
ENSMUST00000175745.2
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr5_+_24890810 | 0.03 |
ENSMUST00000068825.8
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr6_+_41098273 | 0.03 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr12_+_38830812 | 0.02 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr6_+_41107047 | 0.02 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr11_+_101623836 | 0.02 |
ENSMUST00000129741.2
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr9_-_71803354 | 0.02 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr6_-_69394425 | 0.02 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr12_-_115083839 | 0.02 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr2_-_84865831 | 0.02 |
ENSMUST00000028465.14
|
P2rx3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr12_+_38831093 | 0.02 |
ENSMUST00000161513.9
|
Etv1
|
ets variant 1 |
| chr8_+_46080840 | 0.02 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_+_41331039 | 0.01 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr12_-_115299134 | 0.01 |
ENSMUST00000195359.6
ENSMUST00000103530.3 |
Ighv1-59
|
immunoglobulin heavy variable V1-59 |
| chr8_+_88925821 | 0.01 |
ENSMUST00000066748.10
ENSMUST00000119033.8 ENSMUST00000118952.8 |
Tent4b
|
terminal nucleotidyltransferase 4B |
| chr6_-_68784692 | 0.01 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr2_+_154390808 | 0.01 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr2_-_147028309 | 0.01 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr3_+_96604415 | 0.01 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr15_-_76419131 | 0.01 |
ENSMUST00000230604.2
|
Tmem249
|
transmembrane protein 249 |
| chr3_-_92926364 | 0.01 |
ENSMUST00000193944.2
ENSMUST00000029520.9 |
Lce1m
|
late cornified envelope 1M |
| chr6_+_142359099 | 0.01 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr6_+_68402550 | 0.01 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr2_-_86809141 | 0.01 |
ENSMUST00000094913.6
|
Olfr1100
|
olfactory receptor 1100 |
| chr19_-_39637489 | 0.01 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr14_-_52273600 | 0.01 |
ENSMUST00000214342.2
|
Olfr221
|
olfactory receptor 221 |
| chr18_-_3281727 | 0.01 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chr6_+_21986445 | 0.01 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr1_+_100025486 | 0.00 |
ENSMUST00000188735.2
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr6_+_123206880 | 0.00 |
ENSMUST00000205129.2
|
Clec4n
|
C-type lectin domain family 4, member n |
| chrX_-_142716085 | 0.00 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr1_-_28819331 | 0.00 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr17_+_21534225 | 0.00 |
ENSMUST00000077301.6
|
Vmn1r237
|
vomeronasal 1 receptor 237 |
| chr18_-_3281089 | 0.00 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr19_-_39729431 | 0.00 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr17_+_37777099 | 0.00 |
ENSMUST00000031086.5
|
Olfr109
|
olfactory receptor 109 |
| chr13_-_22403990 | 0.00 |
ENSMUST00000057516.2
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr4_+_62443606 | 0.00 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr5_-_137531471 | 0.00 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr2_-_89127612 | 0.00 |
ENSMUST00000099787.2
|
Olfr1230
|
olfactory receptor 1230 |
| chr7_-_80554807 | 0.00 |
ENSMUST00000119428.2
ENSMUST00000026817.11 |
Nmb
|
neuromedin B |
| chr7_-_132414823 | 0.00 |
ENSMUST00000106165.8
|
Fam53b
|
family with sequence similarity 53, member B |
| chr6_+_58242159 | 0.00 |
ENSMUST00000228678.2
|
Vmn1r28
|
vomeronasal 1 receptor 28 |
| chr6_+_70332836 | 0.00 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr14_-_8798841 | 0.00 |
ENSMUST00000061045.3
|
Sntn
|
sentan, cilia apical structure protein |
| chr9_+_19227643 | 0.00 |
ENSMUST00000220268.2
|
Olfr844
|
olfactory receptor 844 |
| chr14_+_26414422 | 0.00 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr8_-_40761372 | 0.00 |
ENSMUST00000118639.2
|
Fgf20
|
fibroblast growth factor 20 |
| chr18_-_3281752 | 0.00 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr17_+_70828649 | 0.00 |
ENSMUST00000233283.2
|
Dlgap1
|
DLG associated protein 1 |
| chr19_-_8196196 | 0.00 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 0.4 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.2 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.5 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.3 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.2 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |