Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
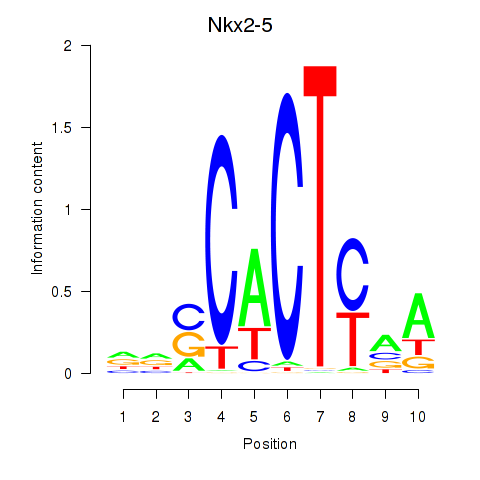
Results for Nkx2-5
Z-value: 1.22
Transcription factors associated with Nkx2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-5
|
ENSMUSG00000015579.6 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-5 | mm39_v1_chr17_-_27060539_27060565 | 0.37 | 5.4e-01 | Click! |
Activity profile of Nkx2-5 motif
Sorted Z-values of Nkx2-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_96868483 | 0.83 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr14_+_51366512 | 0.82 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr16_+_44764099 | 0.80 |
ENSMUST00000114622.10
ENSMUST00000166731.2 |
Cd200r3
|
CD200 receptor 3 |
| chr15_-_82108531 | 0.78 |
ENSMUST00000109535.3
ENSMUST00000089161.10 |
Tnfrsf13c
|
tumor necrosis factor receptor superfamily, member 13c |
| chr4_+_85123654 | 0.72 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr7_+_127845984 | 0.70 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr16_+_43993599 | 0.70 |
ENSMUST00000119746.8
ENSMUST00000088356.10 ENSMUST00000169582.3 |
Usf3
|
upstream transcription factor family member 3 |
| chr3_-_129834788 | 0.70 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr17_+_69071546 | 0.62 |
ENSMUST00000233625.2
|
L3mbtl4
|
L3MBTL4 histone methyl-lysine binding protein |
| chr1_+_74752710 | 0.57 |
ENSMUST00000027356.7
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr14_+_51366306 | 0.57 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr9_+_54824547 | 0.54 |
ENSMUST00000039742.8
|
Hykk
|
hydroxylysine kinase 1 |
| chr11_+_32483290 | 0.53 |
ENSMUST00000102821.4
|
Stk10
|
serine/threonine kinase 10 |
| chr8_+_125519751 | 0.53 |
ENSMUST00000041106.9
ENSMUST00000167588.9 |
Trim67
|
tripartite motif-containing 67 |
| chr15_-_79212323 | 0.48 |
ENSMUST00000166977.9
|
Pla2g6
|
phospholipase A2, group VI |
| chr7_+_28869770 | 0.47 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr9_+_44684248 | 0.41 |
ENSMUST00000128150.8
|
Ift46
|
intraflagellar transport 46 |
| chr19_+_53131187 | 0.40 |
ENSMUST00000050096.15
ENSMUST00000237832.2 |
Add3
|
adducin 3 (gamma) |
| chr15_-_99355623 | 0.34 |
ENSMUST00000023747.14
|
Nckap5l
|
NCK-associated protein 5-like |
| chr15_-_5273645 | 0.34 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr16_+_33504908 | 0.33 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr15_-_5273659 | 0.33 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr14_+_21550921 | 0.33 |
ENSMUST00000182964.3
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr19_-_59931432 | 0.32 |
ENSMUST00000170819.2
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr9_+_44684285 | 0.31 |
ENSMUST00000239014.2
|
Ift46
|
intraflagellar transport 46 |
| chr2_-_73722874 | 0.29 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chr7_+_127846121 | 0.25 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr9_-_53447908 | 0.25 |
ENSMUST00000150244.2
|
Atm
|
ataxia telangiectasia mutated |
| chr9_+_98305014 | 0.24 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chrX_+_164953444 | 0.24 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr4_+_62537750 | 0.24 |
ENSMUST00000084521.11
ENSMUST00000107424.2 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr5_+_16758777 | 0.24 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr7_+_28869629 | 0.24 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr4_-_93223746 | 0.23 |
ENSMUST00000066774.6
|
Tusc1
|
tumor suppressor candidate 1 |
| chr2_+_173918089 | 0.23 |
ENSMUST00000155000.8
ENSMUST00000134876.8 ENSMUST00000147038.8 |
Stx16
|
syntaxin 16 |
| chr6_+_92846338 | 0.22 |
ENSMUST00000113434.2
|
Gm15737
|
predicted gene 15737 |
| chr3_-_107145968 | 0.22 |
ENSMUST00000197758.5
|
Prok1
|
prokineticin 1 |
| chr11_-_101877832 | 0.21 |
ENSMUST00000107173.9
ENSMUST00000107172.8 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr6_+_72281587 | 0.21 |
ENSMUST00000183018.8
ENSMUST00000182014.10 |
Sftpb
|
surfactant associated protein B |
| chr9_+_44684206 | 0.20 |
ENSMUST00000002099.11
|
Ift46
|
intraflagellar transport 46 |
| chr2_-_73722932 | 0.20 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chr6_+_47221293 | 0.20 |
ENSMUST00000199100.5
|
Cntnap2
|
contactin associated protein-like 2 |
| chr5_+_16758897 | 0.19 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chrX_-_63320543 | 0.19 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr8_-_33374282 | 0.19 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr1_-_133589020 | 0.18 |
ENSMUST00000193504.6
ENSMUST00000195067.2 ENSMUST00000191896.6 ENSMUST00000194668.6 ENSMUST00000195424.6 ENSMUST00000179598.4 ENSMUST00000027736.13 |
Zc3h11a
Gm38394
Zc3h11a
|
zinc finger CCCH type containing 11A predicted gene, 38394 zinc finger CCCH type containing 11A |
| chr17_+_27152276 | 0.18 |
ENSMUST00000237412.2
|
Phf1
|
PHD finger protein 1 |
| chr1_+_172168764 | 0.18 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr1_-_66856695 | 0.17 |
ENSMUST00000068168.10
ENSMUST00000113987.2 |
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr5_+_124466146 | 0.17 |
ENSMUST00000111477.2
ENSMUST00000077376.3 |
2810006K23Rik
|
RIKEN cDNA 2810006K23 gene |
| chr12_+_91367764 | 0.16 |
ENSMUST00000021346.14
ENSMUST00000021343.8 |
Tshr
|
thyroid stimulating hormone receptor |
| chr6_-_115830307 | 0.16 |
ENSMUST00000032469.13
|
Mbd4
|
methyl-CpG binding domain protein 4 |
| chr10_+_79824418 | 0.16 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr6_-_29380467 | 0.16 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr5_-_124465875 | 0.15 |
ENSMUST00000184951.8
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr2_-_32977182 | 0.14 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr14_+_20724378 | 0.13 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr7_-_126496534 | 0.13 |
ENSMUST00000120007.8
|
Tmem219
|
transmembrane protein 219 |
| chr6_+_136530970 | 0.12 |
ENSMUST00000189535.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_-_28421678 | 0.12 |
ENSMUST00000090511.4
|
Gcc1
|
golgi coiled coil 1 |
| chr2_+_51928017 | 0.12 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr18_+_49965689 | 0.11 |
ENSMUST00000180611.8
|
Dmxl1
|
Dmx-like 1 |
| chr2_-_73316809 | 0.11 |
ENSMUST00000141264.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_-_89067462 | 0.10 |
ENSMUST00000029686.4
|
Hcn3
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
| chr10_+_22520910 | 0.10 |
ENSMUST00000042261.5
|
Slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr6_-_136899167 | 0.10 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr14_+_80237691 | 0.10 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr6_+_57234937 | 0.09 |
ENSMUST00000228297.2
|
Vmn1r15
|
vomeronasal 1 receptor 15 |
| chr6_+_127554931 | 0.09 |
ENSMUST00000071563.5
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr7_+_43339842 | 0.08 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr9_+_44684450 | 0.07 |
ENSMUST00000238800.2
ENSMUST00000147559.8 |
Ift46
|
intraflagellar transport 46 |
| chr11_+_68858942 | 0.07 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr14_-_63509131 | 0.07 |
ENSMUST00000132122.2
|
Gata4
|
GATA binding protein 4 |
| chr11_-_97909134 | 0.07 |
ENSMUST00000107561.9
|
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr15_-_79212400 | 0.07 |
ENSMUST00000173163.8
ENSMUST00000047816.15 ENSMUST00000172403.9 ENSMUST00000173632.8 |
Pla2g6
|
phospholipase A2, group VI |
| chr11_-_96807192 | 0.06 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr15_-_99355593 | 0.06 |
ENSMUST00000161948.2
|
Nckap5l
|
NCK-associated protein 5-like |
| chr6_-_115830249 | 0.06 |
ENSMUST00000122816.3
|
Mbd4
|
methyl-CpG binding domain protein 4 |
| chr14_+_20724366 | 0.06 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr5_-_124465946 | 0.05 |
ENSMUST00000031344.13
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr11_+_117223161 | 0.05 |
ENSMUST00000106349.2
|
Septin9
|
septin 9 |
| chr12_-_115299134 | 0.05 |
ENSMUST00000195359.6
ENSMUST00000103530.3 |
Ighv1-59
|
immunoglobulin heavy variable V1-59 |
| chr2_+_32498716 | 0.05 |
ENSMUST00000129165.2
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr19_+_16933471 | 0.04 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chrX_-_164110372 | 0.03 |
ENSMUST00000058787.9
|
Glra2
|
glycine receptor, alpha 2 subunit |
| chr11_+_119205613 | 0.03 |
ENSMUST00000053245.7
|
Card14
|
caspase recruitment domain family, member 14 |
| chr8_+_114369838 | 0.03 |
ENSMUST00000095173.3
ENSMUST00000034219.12 ENSMUST00000212269.2 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr4_+_119494901 | 0.03 |
ENSMUST00000024015.3
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr14_-_78326435 | 0.03 |
ENSMUST00000118785.3
ENSMUST00000066437.5 |
Fam216b
|
family with sequence similarity 216, member B |
| chr5_-_8417982 | 0.03 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr18_-_68562385 | 0.03 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr7_-_97794679 | 0.02 |
ENSMUST00000098281.4
|
Omp
|
olfactory marker protein |
| chr12_-_115611981 | 0.02 |
ENSMUST00000103540.3
ENSMUST00000199266.2 |
Ighv8-12
|
immunoglobulin heavy variable V8-12 |
| chrX_-_110372843 | 0.02 |
ENSMUST00000096348.10
ENSMUST00000113428.9 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr6_+_30639217 | 0.02 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr3_+_97808506 | 0.01 |
ENSMUST00000029476.9
ENSMUST00000130620.2 ENSMUST00000122288.2 |
Sec22b
|
SEC22 homolog B, vesicle trafficking protein |
| chr8_-_84301457 | 0.01 |
ENSMUST00000238997.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr6_-_29380423 | 0.01 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chrX_-_110372800 | 0.01 |
ENSMUST00000137712.9
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr14_+_53073315 | 0.01 |
ENSMUST00000197128.2
|
Trav7d-5
|
T cell receptor alpha variable 7D-5 |
| chrX_-_110372733 | 0.01 |
ENSMUST00000065976.12
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr9_+_44684324 | 0.01 |
ENSMUST00000214854.3
ENSMUST00000125877.8 |
Ift46
|
intraflagellar transport 46 |
| chr6_+_30541581 | 0.01 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr11_+_87457479 | 0.01 |
ENSMUST00000239011.2
|
Septin4
|
septin 4 |
| chrX_+_12937714 | 0.01 |
ENSMUST00000169594.9
ENSMUST00000089302.11 |
Usp9x
|
ubiquitin specific peptidase 9, X chromosome |
| chr1_+_74317709 | 0.01 |
ENSMUST00000077985.4
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr9_-_10904714 | 0.01 |
ENSMUST00000162484.8
ENSMUST00000160216.8 |
Cntn5
|
contactin 5 |
| chr11_+_87457544 | 0.01 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr3_+_3699205 | 0.01 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chrX_-_140508177 | 0.00 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr13_-_64645606 | 0.00 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr3_-_57483330 | 0.00 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr5_+_71815382 | 0.00 |
ENSMUST00000199967.5
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr5_-_71815318 | 0.00 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr6_+_14901439 | 0.00 |
ENSMUST00000128567.8
|
Foxp2
|
forkhead box P2 |
| chr12_-_114909863 | 0.00 |
ENSMUST00000103517.2
ENSMUST00000195417.2 |
Ighv1-43
|
immunoglobulin heavy variable V1-43 |
| chr7_-_134100659 | 0.00 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000388 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.2 | 0.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.8 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.5 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.3 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.1 | 0.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 1.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 1.0 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0060464 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.5 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.2 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |