Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
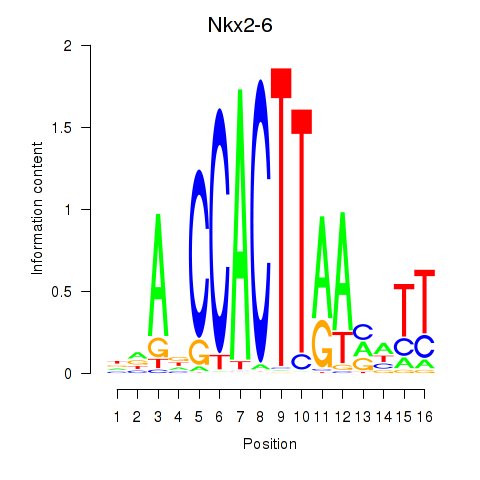
Results for Nkx2-6
Z-value: 0.32
Transcription factors associated with Nkx2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-6
|
ENSMUSG00000044186.11 | NK2 homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-6 | mm39_v1_chr14_+_69409251_69409414 | -0.10 | 8.7e-01 | Click! |
Activity profile of Nkx2-6 motif
Sorted Z-values of Nkx2-6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_8779903 | 0.19 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr15_+_73595012 | 0.14 |
ENSMUST00000230044.2
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr1_+_179788037 | 0.11 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr19_+_55240357 | 0.11 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr5_+_110478558 | 0.09 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr2_-_121287174 | 0.09 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chr3_+_87432879 | 0.09 |
ENSMUST00000170036.8
ENSMUST00000117293.2 |
Etv3
|
ets variant 3 |
| chr1_+_179788675 | 0.07 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr15_+_73594965 | 0.07 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr10_+_4297762 | 0.06 |
ENSMUST00000215696.2
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr9_-_102496047 | 0.05 |
ENSMUST00000215253.2
|
Cep63
|
centrosomal protein 63 |
| chr15_-_78657640 | 0.05 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_22959223 | 0.04 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr2_+_22959452 | 0.04 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr17_-_45910529 | 0.04 |
ENSMUST00000171847.8
ENSMUST00000166633.8 ENSMUST00000169729.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr6_-_41681273 | 0.03 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr4_-_109522502 | 0.02 |
ENSMUST00000063531.5
|
Cdkn2c
|
cyclin dependent kinase inhibitor 2C |
| chr7_-_115837034 | 0.01 |
ENSMUST00000182834.8
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr12_-_114263874 | 0.01 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr12_-_114073050 | 0.01 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr16_+_34815177 | 0.01 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr14_-_73286535 | 0.01 |
ENSMUST00000169168.3
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr2_+_3115250 | 0.00 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr14_-_73286504 | 0.00 |
ENSMUST00000044664.12
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr16_-_58647137 | 0.00 |
ENSMUST00000215069.2
ENSMUST00000079955.6 |
Olfr175
|
olfactory receptor 175 |
| chr2_-_147028309 | 0.00 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr7_-_107855343 | 0.00 |
ENSMUST00000072968.2
|
Olfr488
|
olfactory receptor 488 |
| chr2_-_119060366 | 0.00 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr15_-_37734579 | 0.00 |
ENSMUST00000145909.9
ENSMUST00000153775.9 |
Gm49397
Ncald
|
predicted gene, 49397 neurocalcin delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.0 | 0.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |