Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
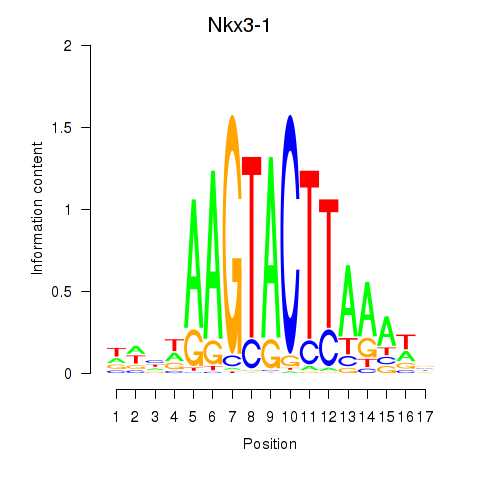
Results for Nkx3-1
Z-value: 0.92
Transcription factors associated with Nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-1
|
ENSMUSG00000022061.9 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx3-1 | mm39_v1_chr14_+_69428087_69428140 | 0.88 | 4.9e-02 | Click! |
Activity profile of Nkx3-1 motif
Sorted Z-values of Nkx3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23940964 | 1.30 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr13_+_23755551 | 1.06 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr17_-_57289121 | 0.70 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chr16_-_48592372 | 0.67 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr9_-_43027809 | 0.66 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr9_-_39863342 | 0.63 |
ENSMUST00000216647.2
|
Olfr975
|
olfactory receptor 975 |
| chr11_-_69649004 | 0.54 |
ENSMUST00000071213.4
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr17_-_66079629 | 0.52 |
ENSMUST00000233258.2
|
Rab31
|
RAB31, member RAS oncogene family |
| chr11_+_68989763 | 0.50 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr19_-_12147438 | 0.48 |
ENSMUST00000207679.3
ENSMUST00000219261.2 |
Olfr1555-ps1
|
olfactory receptor 1555, pseudogene 1 |
| chr11_+_45946800 | 0.47 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr19_+_13608985 | 0.47 |
ENSMUST00000217182.3
|
Olfr1489
|
olfactory receptor 1489 |
| chr4_-_15149755 | 0.47 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr12_+_76371634 | 0.46 |
ENSMUST00000154078.3
ENSMUST00000095610.9 |
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr16_-_58695131 | 0.45 |
ENSMUST00000217377.2
|
Olfr177
|
olfactory receptor 177 |
| chr2_-_88534814 | 0.41 |
ENSMUST00000216928.2
ENSMUST00000216977.2 |
Olfr1196
|
olfactory receptor 1196 |
| chr5_-_86824205 | 0.40 |
ENSMUST00000038448.7
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chr7_+_107649900 | 0.39 |
ENSMUST00000214599.2
ENSMUST00000209805.3 |
Olfr479
|
olfactory receptor 479 |
| chr2_-_89951611 | 0.38 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr19_+_43678109 | 0.37 |
ENSMUST00000081079.6
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr10_-_4382311 | 0.37 |
ENSMUST00000126102.8
ENSMUST00000131853.2 ENSMUST00000042251.11 |
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr1_-_173569301 | 0.36 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr1_+_174251630 | 0.36 |
ENSMUST00000192358.3
|
Olfr414
|
olfactory receptor 414 |
| chr2_-_87838612 | 0.36 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr2_-_85966272 | 0.35 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr15_-_74983786 | 0.34 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr10_+_129539079 | 0.34 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804 |
| chr8_-_123405392 | 0.34 |
ENSMUST00000134045.2
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr7_-_28297565 | 0.34 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr8_-_96534043 | 0.33 |
ENSMUST00000213046.2
|
Cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr10_-_13199971 | 0.33 |
ENSMUST00000105543.9
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr12_+_102249294 | 0.32 |
ENSMUST00000056950.14
|
Rin3
|
Ras and Rab interactor 3 |
| chr9_-_88974735 | 0.32 |
ENSMUST00000189557.2
ENSMUST00000167113.8 |
Trim43b
|
tripartite motif-containing 43B |
| chr18_+_75151780 | 0.32 |
ENSMUST00000236220.2
ENSMUST00000039608.9 ENSMUST00000235692.2 |
Dym
|
dymeclin |
| chr19_-_59931432 | 0.25 |
ENSMUST00000170819.2
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr2_-_89408791 | 0.25 |
ENSMUST00000217402.2
|
Olfr1245
|
olfactory receptor 1245 |
| chr12_-_100865783 | 0.25 |
ENSMUST00000053668.10
|
Gpr68
|
G protein-coupled receptor 68 |
| chr6_+_89946295 | 0.25 |
ENSMUST00000205088.3
ENSMUST00000226715.2 ENSMUST00000228401.2 |
Vmn1r46
|
vomeronasal 1 receptor 46 |
| chr7_+_5221492 | 0.23 |
ENSMUST00000228062.2
ENSMUST00000227798.2 |
Vmn1r57
|
vomeronasal 1 receptor 57 |
| chr4_-_120874376 | 0.23 |
ENSMUST00000043200.8
|
Smap2
|
small ArfGAP 2 |
| chr13_-_42000958 | 0.22 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_+_35354148 | 0.20 |
ENSMUST00000166426.9
ENSMUST00000025250.14 |
Bag6
|
BCL2-associated athanogene 6 |
| chr2_+_89842475 | 0.20 |
ENSMUST00000214382.2
ENSMUST00000217065.3 |
Olfr1263
|
olfactory receptor 1263 |
| chr9_+_72345801 | 0.20 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr16_+_38279289 | 0.20 |
ENSMUST00000099816.3
ENSMUST00000232409.2 |
Cd80
|
CD80 antigen |
| chr17_+_37710293 | 0.20 |
ENSMUST00000216844.2
ENSMUST00000215974.2 ENSMUST00000215894.2 |
Olfr107
|
olfactory receptor 107 |
| chr3_-_64417263 | 0.19 |
ENSMUST00000177184.9
|
Vmn2r5
|
vomeronasal 2, receptor 5 |
| chr7_-_24937276 | 0.19 |
ENSMUST00000071739.12
ENSMUST00000108411.2 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr3_-_95595157 | 0.19 |
ENSMUST00000015994.4
ENSMUST00000148854.2 ENSMUST00000117782.8 |
Adamtsl4
|
ADAMTS-like 4 |
| chr2_-_111908072 | 0.19 |
ENSMUST00000213577.2
ENSMUST00000216071.2 |
Olfr1313
|
olfactory receptor 1313 |
| chr10_-_14593935 | 0.19 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr17_+_43327412 | 0.19 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr12_-_77008799 | 0.19 |
ENSMUST00000218640.2
|
Max
|
Max protein |
| chr15_-_50746202 | 0.18 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr11_+_101169918 | 0.18 |
ENSMUST00000103107.5
|
Cntd1
|
cyclin N-terminal domain containing 1 |
| chr11_+_74077075 | 0.18 |
ENSMUST00000206114.3
|
Olfr403
|
olfactory receptor 403 |
| chr19_-_17333972 | 0.18 |
ENSMUST00000174236.8
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr16_-_11727262 | 0.17 |
ENSMUST00000127972.8
ENSMUST00000121750.2 ENSMUST00000096272.11 ENSMUST00000073371.7 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr8_+_114860342 | 0.17 |
ENSMUST00000109109.8
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_-_96534085 | 0.16 |
ENSMUST00000098473.11
ENSMUST00000211887.2 |
Cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr17_+_35354172 | 0.16 |
ENSMUST00000172571.8
ENSMUST00000173491.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr17_+_38456172 | 0.16 |
ENSMUST00000215078.3
ENSMUST00000215549.3 ENSMUST00000173610.2 |
Olfr133
|
olfactory receptor 133 |
| chr7_+_75105282 | 0.16 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr7_+_135207505 | 0.16 |
ENSMUST00000210833.2
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr2_+_37029334 | 0.15 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chr1_-_155910567 | 0.15 |
ENSMUST00000141878.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr2_+_79538124 | 0.15 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr8_-_3798941 | 0.15 |
ENSMUST00000012847.3
|
Cd209a
|
CD209a antigen |
| chr1_-_80642969 | 0.14 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr6_+_137229332 | 0.14 |
ENSMUST00000167679.8
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr11_-_73800125 | 0.14 |
ENSMUST00000215690.2
|
Olfr395
|
olfactory receptor 395 |
| chr15_+_98065039 | 0.14 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr8_-_3798922 | 0.14 |
ENSMUST00000208960.2
ENSMUST00000207979.2 |
Cd209a
|
CD209a antigen |
| chr7_-_85610837 | 0.14 |
ENSMUST00000232886.2
ENSMUST00000232758.2 ENSMUST00000166355.3 |
Vmn2r74
|
vomeronasal 2, receptor 74 |
| chr7_+_29467971 | 0.14 |
ENSMUST00000032802.5
|
Zfp84
|
zinc finger protein 84 |
| chr4_+_62537750 | 0.13 |
ENSMUST00000084521.11
ENSMUST00000107424.2 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr6_-_41012435 | 0.13 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr10_+_25317309 | 0.13 |
ENSMUST00000217929.2
ENSMUST00000220121.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr2_-_88810595 | 0.13 |
ENSMUST00000168169.10
ENSMUST00000144908.3 |
Olfr1213
|
olfactory receptor 1213 |
| chr10_-_129751466 | 0.12 |
ENSMUST00000213438.2
|
Olfr816
|
olfactory receptor 816 |
| chr19_-_12209960 | 0.12 |
ENSMUST00000207710.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr1_-_133834790 | 0.12 |
ENSMUST00000149380.8
ENSMUST00000124051.9 |
Optc
|
opticin |
| chr7_-_43906802 | 0.12 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr9_+_20209828 | 0.12 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr12_-_115832846 | 0.11 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr1_-_155910546 | 0.11 |
ENSMUST00000169241.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr3_-_20296337 | 0.11 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr18_+_61096597 | 0.11 |
ENSMUST00000115295.9
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr10_+_78848306 | 0.10 |
ENSMUST00000216030.2
|
Olfr1351
|
olfactory receptor 1351 |
| chr4_-_140501507 | 0.10 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr3_-_113166153 | 0.10 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr3_+_96603694 | 0.10 |
ENSMUST00000107076.10
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr19_+_11446716 | 0.09 |
ENSMUST00000165310.3
|
Ms4a6c
|
membrane-spanning 4-domains, subfamily A, member 6C |
| chr6_+_54016543 | 0.09 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr1_-_52992330 | 0.09 |
ENSMUST00000114492.8
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr16_+_56024676 | 0.09 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr19_-_10217968 | 0.09 |
ENSMUST00000189897.2
ENSMUST00000186056.7 ENSMUST00000088013.12 |
Myrf
|
myelin regulatory factor |
| chr6_-_128339775 | 0.09 |
ENSMUST00000112152.8
ENSMUST00000057421.15 ENSMUST00000112151.2 |
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr9_+_38516398 | 0.08 |
ENSMUST00000217057.2
|
Olfr914
|
olfactory receptor 914 |
| chr13_+_109769294 | 0.08 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr12_-_98700886 | 0.08 |
ENSMUST00000085116.4
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr5_-_69749617 | 0.08 |
ENSMUST00000173927.8
ENSMUST00000120789.8 ENSMUST00000031117.13 |
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr3_-_113263974 | 0.08 |
ENSMUST00000098667.5
|
Amy2a2
|
amylase 2a2 |
| chr17_-_35381945 | 0.08 |
ENSMUST00000174805.2
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr6_+_68279392 | 0.07 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr7_-_107857554 | 0.07 |
ENSMUST00000215173.2
|
Olfr488
|
olfactory receptor 488 |
| chr7_-_5808444 | 0.07 |
ENSMUST00000075085.7
|
Vmn1r63
|
vomeronasal 1 receptor 63 |
| chr3_-_113231368 | 0.07 |
ENSMUST00000179314.3
|
Amy2a3
|
amylase 2a3 |
| chr19_-_8382424 | 0.07 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr1_-_80643024 | 0.06 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr2_-_57014015 | 0.06 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_-_121410152 | 0.06 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr11_-_101169753 | 0.06 |
ENSMUST00000168089.2
ENSMUST00000017332.4 |
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chr7_+_110226857 | 0.06 |
ENSMUST00000033054.10
|
Adm
|
adrenomedullin |
| chr2_+_87078357 | 0.06 |
ENSMUST00000214119.2
ENSMUST00000217196.3 ENSMUST00000213513.2 |
Olfr1115
|
olfactory receptor 1115 |
| chr9_+_74883377 | 0.06 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr11_+_95152355 | 0.06 |
ENSMUST00000021242.5
|
Tac4
|
tachykinin 4 |
| chr3_-_113198765 | 0.06 |
ENSMUST00000179568.3
|
Amy2a4
|
amylase 2a4 |
| chr17_-_80022480 | 0.05 |
ENSMUST00000234361.2
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr3_-_127202586 | 0.05 |
ENSMUST00000183095.3
ENSMUST00000182610.8 |
Ank2
|
ankyrin 2, brain |
| chr1_-_69723316 | 0.05 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr7_-_10011933 | 0.05 |
ENSMUST00000227719.2
ENSMUST00000228622.2 ENSMUST00000228086.2 |
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr14_+_51599213 | 0.05 |
ENSMUST00000162998.2
|
Gm7247
|
predicted gene 7247 |
| chr8_+_85952617 | 0.05 |
ENSMUST00000218663.2
|
Olfr371
|
olfactory receptor 371 |
| chr5_-_134776101 | 0.04 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr3_-_127202693 | 0.04 |
ENSMUST00000182078.9
|
Ank2
|
ankyrin 2, brain |
| chr1_+_172168764 | 0.04 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr14_-_55101505 | 0.04 |
ENSMUST00000142283.4
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr12_+_76371679 | 0.04 |
ENSMUST00000172992.2
|
Akap5
|
A kinase (PRKA) anchor protein 5 |
| chr17_-_48003391 | 0.04 |
ENSMUST00000113300.8
|
Prickle4
|
prickle planar cell polarity protein 4 |
| chr17_-_15784582 | 0.04 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chrY_+_57195716 | 0.03 |
ENSMUST00000189109.7
ENSMUST00000191355.7 ENSMUST00000190292.2 |
Sly
|
Sycp3 like Y-linked |
| chr3_-_127202663 | 0.03 |
ENSMUST00000182008.8
ENSMUST00000182547.8 |
Ank2
|
ankyrin 2, brain |
| chr14_+_61547202 | 0.03 |
ENSMUST00000055159.8
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr11_-_73290321 | 0.03 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr5_-_21629661 | 0.03 |
ENSMUST00000115245.8
ENSMUST00000030552.7 |
Ccdc146
|
coiled-coil domain containing 146 |
| chr4_-_108075119 | 0.03 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr12_-_103660916 | 0.03 |
ENSMUST00000117053.8
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr4_+_110254858 | 0.02 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chrX_-_100266032 | 0.02 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr8_-_3722290 | 0.02 |
ENSMUST00000159364.3
|
Fcor
|
Foxo1 corepressor |
| chr7_+_138794577 | 0.02 |
ENSMUST00000135509.2
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr1_-_192718064 | 0.02 |
ENSMUST00000215093.2
ENSMUST00000195354.6 |
Syt14
|
synaptotagmin XIV |
| chr8_+_124059414 | 0.02 |
ENSMUST00000010298.7
|
Spire2
|
spire type actin nucleation factor 2 |
| chr15_+_100320028 | 0.02 |
ENSMUST00000132119.2
|
Gm5475
|
predicted gene 5475 |
| chr2_-_89084074 | 0.02 |
ENSMUST00000216392.2
|
Olfr1228
|
olfactory receptor 1228 |
| chr13_+_35059285 | 0.02 |
ENSMUST00000077853.5
|
Prpf4b
|
pre-mRNA processing factor 4B |
| chr6_+_86055018 | 0.02 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr2_-_84605732 | 0.02 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr3_-_87934772 | 0.02 |
ENSMUST00000005014.9
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chrX_-_99669507 | 0.02 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr19_+_13037974 | 0.02 |
ENSMUST00000213806.2
ENSMUST00000214695.2 ENSMUST00000217568.2 |
Olfr1454
|
olfactory receptor 1454 |
| chr9_+_77829191 | 0.02 |
ENSMUST00000133757.8
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr6_+_86055048 | 0.02 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta) |
| chr14_-_21798678 | 0.01 |
ENSMUST00000075040.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr9_+_37765565 | 0.01 |
ENSMUST00000213956.2
|
Olfr877
|
olfactory receptor 877 |
| chrX_+_163052367 | 0.01 |
ENSMUST00000145412.8
ENSMUST00000033749.9 |
Pir
|
pirin |
| chr4_+_110254907 | 0.01 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr17_-_37613523 | 0.01 |
ENSMUST00000215392.2
|
Olfr101
|
olfactory receptor 101 |
| chr19_-_13074935 | 0.01 |
ENSMUST00000214561.3
|
Olfr1457
|
olfactory receptor 1457 |
| chr1_-_173594475 | 0.01 |
ENSMUST00000111214.4
|
Ifi204
|
interferon activated gene 204 |
| chrX_-_74918122 | 0.01 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr1_-_86317066 | 0.01 |
ENSMUST00000212058.2
|
Nmur1
|
neuromedin U receptor 1 |
| chrY_+_65386218 | 0.01 |
ENSMUST00000190282.7
ENSMUST00000185550.7 |
Gm20736
|
predicted gene, 20736 |
| chrY_+_6583546 | 0.01 |
ENSMUST00000190026.2
|
Gm28919
|
predicted gene 28919 |
| chr14_+_61547267 | 0.01 |
ENSMUST00000224727.2
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chrY_-_40272248 | 0.01 |
ENSMUST00000191443.7
|
Gm21865
|
predicted gene, 21865 |
| chr1_-_162694076 | 0.01 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_+_132688558 | 0.01 |
ENSMUST00000028835.13
ENSMUST00000110122.10 |
Crls1
|
cardiolipin synthase 1 |
| chr7_-_108471586 | 0.01 |
ENSMUST00000216500.2
|
Olfr517
|
olfactory receptor 517 |
| chr7_-_106433100 | 0.01 |
ENSMUST00000219803.2
ENSMUST00000208864.2 |
Olfr702
|
olfactory receptor 702 |
| chr15_-_11907267 | 0.01 |
ENSMUST00000228489.2
|
Npr3
|
natriuretic peptide receptor 3 |
| chrY_+_57714225 | 0.01 |
ENSMUST00000190573.2
|
Gm28961
|
predicted gene 28961 |
| chr1_+_160970276 | 0.01 |
ENSMUST00000111608.8
ENSMUST00000052245.9 |
Ankrd45
|
ankyrin repeat domain 45 |
| chr6_-_69626340 | 0.01 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chr7_+_126844359 | 0.01 |
ENSMUST00000205583.2
|
Zfp771
|
zinc finger protein 771 |
| chr1_+_170983081 | 0.01 |
ENSMUST00000111334.2
|
Mpz
|
myelin protein zero |
| chrX_+_7786061 | 0.01 |
ENSMUST00000041096.4
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chrY_+_55728335 | 0.01 |
ENSMUST00000185713.7
|
Gm21858
|
predicted gene, 21858 |
| chr12_+_101370932 | 0.01 |
ENSMUST00000055156.5
|
Catsperb
|
cation channel sperm associated auxiliary subunit beta |
| chr7_+_15484302 | 0.01 |
ENSMUST00000098802.10
ENSMUST00000173053.2 |
Obox5
|
oocyte specific homeobox 5 |
| chr5_-_72716942 | 0.01 |
ENSMUST00000074948.5
ENSMUST00000087216.12 |
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr4_-_137137088 | 0.01 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr9_-_40873629 | 0.01 |
ENSMUST00000160120.2
|
Jhy
|
junctional cadherin complex regulator |
| chrY_-_68306157 | 0.01 |
ENSMUST00000189084.7
ENSMUST00000189422.7 |
Gm20937
|
predicted gene, 20937 |
| chrY_+_13252391 | 0.01 |
ENSMUST00000192749.2
ENSMUST00000178599.7 |
Gm21440
Gm21454
|
predicted gene, 21440 predicted gene, 21454 |
| chrY_+_51121799 | 0.01 |
ENSMUST00000185327.7
|
Gm21117
|
predicted gene, 21117 |
| chr4_+_112470794 | 0.01 |
ENSMUST00000058791.14
ENSMUST00000186969.2 |
Skint2
|
selection and upkeep of intraepithelial T cells 2 |
| chrY_-_42537595 | 0.01 |
ENSMUST00000190453.2
|
Gm29276
|
predicted gene 29276 |
| chrY_+_14400710 | 0.01 |
ENSMUST00000192683.2
|
Gm30737
|
predicted gene, 30737 |
| chr9_+_40785277 | 0.01 |
ENSMUST00000067375.5
|
Bsx
|
brain specific homeobox |
| chr7_-_133203838 | 0.01 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chrY_+_19113007 | 0.01 |
ENSMUST00000194086.2
|
Gm35134
|
predicted gene, 35134 |
| chr7_+_102834996 | 0.01 |
ENSMUST00000218618.2
|
Olfr592
|
olfactory receptor 592 |
| chr11_-_45845992 | 0.01 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chrY_+_17874741 | 0.01 |
ENSMUST00000188928.3
ENSMUST00000177639.7 |
Gm20772
Gm20831
|
predicted gene, 20772 predicted gene, 20831 |
| chr6_-_38101503 | 0.01 |
ENSMUST00000040259.8
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr7_+_58307930 | 0.00 |
ENSMUST00000168747.3
|
Atp10a
|
ATPase, class V, type 10A |
| chr7_-_103799746 | 0.00 |
ENSMUST00000059121.5
|
Ubqlnl
|
ubiquilin-like |
| chr15_-_103929556 | 0.00 |
ENSMUST00000226584.2
ENSMUST00000037685.9 |
Mucl2
|
mucin-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.5 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.1 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.4 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0071879 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.4 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.5 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0086023 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.0 | GO:0071603 | trabecular meshwork development(GO:0002930) endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.0 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |