Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
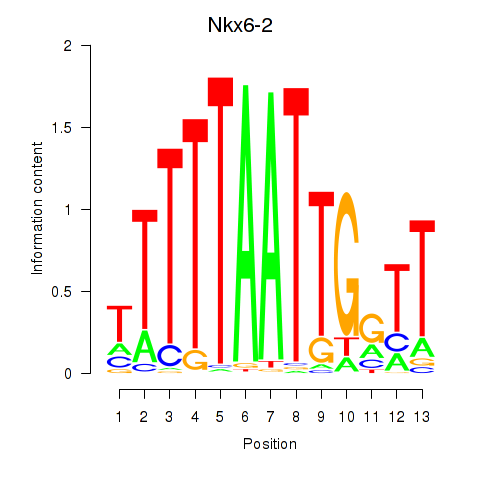
Results for Nkx6-2
Z-value: 0.52
Transcription factors associated with Nkx6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-2
|
ENSMUSG00000041309.18 | NK6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-2 | mm39_v1_chr7_-_139162706_139162724 | 0.16 | 7.9e-01 | Click! |
Activity profile of Nkx6-2 motif
Sorted Z-values of Nkx6-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_154721288 | 0.37 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr3_+_5815863 | 0.31 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chrM_+_9459 | 0.29 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_-_73215442 | 0.24 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr1_-_72251466 | 0.23 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr19_+_32597379 | 0.21 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr4_-_21767116 | 0.18 |
ENSMUST00000029915.6
|
Tstd3
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 3 |
| chr5_-_110987604 | 0.17 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr2_-_174188505 | 0.15 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr14_+_80237691 | 0.14 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr1_+_88062508 | 0.13 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr10_+_19497740 | 0.13 |
ENSMUST00000036564.8
|
Il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr2_+_170353338 | 0.13 |
ENSMUST00000136839.2
ENSMUST00000109148.8 ENSMUST00000170167.8 |
Pfdn4
|
prefoldin 4 |
| chr9_+_51958453 | 0.12 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr8_+_95259618 | 0.12 |
ENSMUST00000048653.10
|
Cpne2
|
copine II |
| chr1_-_183766195 | 0.12 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr14_-_31503869 | 0.11 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr18_-_74340885 | 0.11 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr18_-_74340842 | 0.11 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr2_+_157209506 | 0.11 |
ENSMUST00000081202.6
|
Manbal
|
mannosidase, beta A, lysosomal-like |
| chr18_-_43610829 | 0.11 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_+_69729340 | 0.11 |
ENSMUST00000133967.8
ENSMUST00000094065.5 |
Tmem256
|
transmembrane protein 256 |
| chr19_+_37425180 | 0.11 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr11_+_100902572 | 0.10 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr8_+_95259630 | 0.10 |
ENSMUST00000109537.2
|
Cpne2
|
copine II |
| chrM_+_10167 | 0.10 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_+_74425990 | 0.10 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr16_-_75563645 | 0.10 |
ENSMUST00000114244.2
ENSMUST00000046283.16 |
Hspa13
|
heat shock protein 70 family, member 13 |
| chr15_-_79718423 | 0.09 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr14_+_26722319 | 0.09 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr10_+_79746690 | 0.09 |
ENSMUST00000181321.2
|
Gm26602
|
predicted gene, 26602 |
| chr10_-_127147609 | 0.09 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr3_+_106020545 | 0.09 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr3_-_96128196 | 0.09 |
ENSMUST00000090782.4
|
H2ac20
|
H2A clustered histone 20 |
| chr5_-_122959321 | 0.08 |
ENSMUST00000197074.5
ENSMUST00000199406.5 ENSMUST00000196640.5 ENSMUST00000197719.5 ENSMUST00000200645.5 |
Anapc5
|
anaphase-promoting complex subunit 5 |
| chrX_+_138464065 | 0.08 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr1_+_43484895 | 0.08 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chrX_-_50106844 | 0.08 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr17_-_78991691 | 0.07 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr2_-_131021905 | 0.07 |
ENSMUST00000089510.5
|
Cenpb
|
centromere protein B |
| chr19_-_33728759 | 0.07 |
ENSMUST00000147153.4
|
Lipo2
|
lipase, member O2 |
| chr10_+_88036947 | 0.07 |
ENSMUST00000020248.16
ENSMUST00000182183.8 ENSMUST00000171151.9 ENSMUST00000182619.2 |
Washc3
|
WASH complex subunit 3 |
| chr4_+_147106307 | 0.07 |
ENSMUST00000075775.6
|
Rex2
|
reduced expression 2 |
| chr11_+_62770275 | 0.07 |
ENSMUST00000014321.5
|
Tvp23b
|
trans-golgi network vesicle protein 23B |
| chr18_+_9707595 | 0.07 |
ENSMUST00000234965.2
|
Colec12
|
collectin sub-family member 12 |
| chr1_-_135513443 | 0.07 |
ENSMUST00000067414.13
|
Nav1
|
neuron navigator 1 |
| chr7_+_28488380 | 0.07 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr4_+_147576874 | 0.07 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr3_+_82962823 | 0.07 |
ENSMUST00000150268.8
ENSMUST00000122128.2 |
Plrg1
|
pleiotropic regulator 1 |
| chr10_+_26698556 | 0.06 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr5_+_20112500 | 0.06 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_+_100750316 | 0.06 |
ENSMUST00000107356.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr13_+_110063364 | 0.06 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr4_+_146093394 | 0.06 |
ENSMUST00000168483.9
|
Zfp600
|
zinc finger protein 600 |
| chr9_-_22028419 | 0.05 |
ENSMUST00000214394.2
ENSMUST00000013966.8 |
Elof1
|
ELF1 homolog, elongation factor 1 |
| chr3_+_96128427 | 0.05 |
ENSMUST00000090781.8
|
H2bc21
|
H2B clustered histone 21 |
| chr4_+_146033882 | 0.05 |
ENSMUST00000105730.2
ENSMUST00000091878.6 |
Zfp987
|
zinc finger protein 987 |
| chr4_-_117039809 | 0.05 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr15_+_65682066 | 0.05 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr14_+_51366306 | 0.05 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr5_-_110987441 | 0.05 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr16_-_21980200 | 0.05 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr4_+_145397238 | 0.05 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr11_+_52251687 | 0.05 |
ENSMUST00000102758.8
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr19_-_33495180 | 0.05 |
ENSMUST00000143522.9
|
Lipo4
|
lipase, member O4 |
| chr2_-_69619864 | 0.04 |
ENSMUST00000094942.4
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr1_-_149836974 | 0.04 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr1_+_66360865 | 0.04 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr5_+_140404997 | 0.04 |
ENSMUST00000100507.8
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr4_+_150938376 | 0.04 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr7_-_84328553 | 0.04 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr2_+_163535925 | 0.04 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr5_+_110988095 | 0.04 |
ENSMUST00000198373.2
|
Chek2
|
checkpoint kinase 2 |
| chr9_+_96141299 | 0.04 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr4_+_3940747 | 0.04 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_-_150087587 | 0.04 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr12_-_80807454 | 0.04 |
ENSMUST00000073251.8
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr19_-_44095840 | 0.04 |
ENSMUST00000119591.2
ENSMUST00000026217.11 |
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chrX_-_74425897 | 0.04 |
ENSMUST00000151772.2
ENSMUST00000033539.13 |
F8
|
coagulation factor VIII |
| chr5_+_110987839 | 0.04 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr11_+_100750177 | 0.04 |
ENSMUST00000004145.14
ENSMUST00000133036.8 |
Stat5a
|
signal transducer and activator of transcription 5A |
| chr7_+_44711853 | 0.04 |
ENSMUST00000107829.9
ENSMUST00000003513.11 ENSMUST00000211465.2 ENSMUST00000210088.2 ENSMUST00000210520.2 |
Nosip
|
nitric oxide synthase interacting protein |
| chr6_+_40619913 | 0.04 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr4_+_146586445 | 0.04 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr19_-_33602652 | 0.04 |
ENSMUST00000124230.3
|
Gm8978
|
predicted gene 8978 |
| chr4_+_145241454 | 0.04 |
ENSMUST00000105741.2
|
Zfp990
|
zinc finger protein 990 |
| chr18_-_32044877 | 0.04 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr8_-_62576140 | 0.04 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr11_+_23206001 | 0.03 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr16_-_88759753 | 0.03 |
ENSMUST00000179707.3
|
Krtap16-3
|
keratin associated protein 16-3 |
| chr5_+_66833434 | 0.03 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr19_-_12742811 | 0.03 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr3_-_135373560 | 0.03 |
ENSMUST00000164430.7
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr2_+_69691906 | 0.03 |
ENSMUST00000090852.11
ENSMUST00000166411.8 |
Ssb
|
Sjogren syndrome antigen B |
| chr3_+_68479578 | 0.03 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr3_+_85946145 | 0.03 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr14_+_29700319 | 0.03 |
ENSMUST00000224797.2
|
Actr8
|
ARP8 actin-related protein 8 |
| chr9_-_56151334 | 0.03 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr5_+_92957231 | 0.03 |
ENSMUST00000113054.9
|
Shroom3
|
shroom family member 3 |
| chr11_-_73483834 | 0.03 |
ENSMUST00000215689.2
|
Olfr385
|
olfactory receptor 385 |
| chr7_+_89780785 | 0.03 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr16_-_16647139 | 0.03 |
ENSMUST00000023468.6
|
Spag6l
|
sperm associated antigen 6-like |
| chr14_-_32110312 | 0.03 |
ENSMUST00000100723.4
|
1700024G13Rik
|
RIKEN cDNA 1700024G13 gene |
| chr7_-_19449319 | 0.03 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr3_-_26386609 | 0.03 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1 |
| chr10_+_129691236 | 0.03 |
ENSMUST00000204622.3
|
Olfr813
|
olfactory receptor 813 |
| chr6_-_48063464 | 0.02 |
ENSMUST00000073124.9
ENSMUST00000203609.3 |
Zfp746
|
zinc finger protein 746 |
| chr16_+_44215136 | 0.02 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr8_+_94537910 | 0.02 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr5_+_75312939 | 0.02 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr9_+_96141317 | 0.02 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr4_+_147445744 | 0.02 |
ENSMUST00000133078.8
ENSMUST00000154154.2 |
Zfp978
|
zinc finger protein 978 |
| chr1_-_135513083 | 0.02 |
ENSMUST00000040599.15
|
Nav1
|
neuron navigator 1 |
| chr3_+_96508400 | 0.02 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like |
| chr11_-_109886569 | 0.02 |
ENSMUST00000106669.3
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr9_-_113537277 | 0.02 |
ENSMUST00000111861.4
ENSMUST00000035086.13 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr19_-_17350200 | 0.02 |
ENSMUST00000236139.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr3_+_108000425 | 0.02 |
ENSMUST00000151326.8
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr4_+_145595364 | 0.02 |
ENSMUST00000123460.2
|
Zfp986
|
zinc finger protein 986 |
| chrX_+_110801086 | 0.02 |
ENSMUST00000207962.2
|
Gm45194
|
predicted gene 45194 |
| chr17_+_36172210 | 0.02 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr4_+_45848918 | 0.02 |
ENSMUST00000030011.6
|
Stra6l
|
STRA6-like |
| chr14_+_29700294 | 0.02 |
ENSMUST00000016115.6
|
Actr8
|
ARP8 actin-related protein 8 |
| chr18_+_34973605 | 0.02 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr6_-_13839914 | 0.02 |
ENSMUST00000060442.14
|
Gpr85
|
G protein-coupled receptor 85 |
| chr15_-_13173736 | 0.02 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr10_+_18283405 | 0.02 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr8_-_62355690 | 0.02 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr1_+_66361252 | 0.02 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr2_+_69050315 | 0.02 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr11_+_67212464 | 0.02 |
ENSMUST00000108684.8
|
Myh13
|
myosin, heavy polypeptide 13, skeletal muscle |
| chr10_+_33733706 | 0.02 |
ENSMUST00000218204.2
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr9_-_123507847 | 0.02 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr11_+_73262072 | 0.02 |
ENSMUST00000078952.9
ENSMUST00000120401.9 ENSMUST00000170592.4 |
Olfr376
|
olfactory receptor 376 |
| chr9_-_39427105 | 0.02 |
ENSMUST00000216177.2
|
Olfr957
|
olfactory receptor 957 |
| chr7_+_104583447 | 0.02 |
ENSMUST00000214260.2
ENSMUST00000210138.3 |
Olfr669
|
olfactory receptor 669 |
| chr19_-_33764859 | 0.02 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr5_-_86780277 | 0.02 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr7_-_104854362 | 0.02 |
ENSMUST00000216613.2
|
Olfr686
|
olfactory receptor 686 |
| chr4_-_97472844 | 0.02 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr5_-_87739442 | 0.02 |
ENSMUST00000031201.9
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr5_+_92831468 | 0.02 |
ENSMUST00000168878.8
|
Shroom3
|
shroom family member 3 |
| chr8_-_39128662 | 0.02 |
ENSMUST00000118896.2
|
Sgcz
|
sarcoglycan zeta |
| chr1_-_126758520 | 0.01 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr4_-_147726953 | 0.01 |
ENSMUST00000133006.2
ENSMUST00000037565.14 ENSMUST00000105720.8 |
Zfp979
|
zinc finger protein 979 |
| chr14_-_67246282 | 0.01 |
ENSMUST00000111115.8
ENSMUST00000022634.9 |
Bnip3l
|
BCL2/adenovirus E1B interacting protein 3-like |
| chr8_-_49008305 | 0.01 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr14_-_30213408 | 0.01 |
ENSMUST00000112250.6
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chrX_+_37405054 | 0.01 |
ENSMUST00000016471.9
ENSMUST00000115134.2 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr9_+_39622649 | 0.01 |
ENSMUST00000215164.3
ENSMUST00000213335.2 |
Olfr965
|
olfactory receptor 965 |
| chr5_+_20112771 | 0.01 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_+_65504067 | 0.01 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr7_+_86109129 | 0.01 |
ENSMUST00000217253.2
|
Olfr299
|
olfactory receptor 299 |
| chr14_+_27598021 | 0.01 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr13_+_31740117 | 0.01 |
ENSMUST00000042118.11
|
Foxq1
|
forkhead box Q1 |
| chr2_+_24043159 | 0.01 |
ENSMUST00000028363.2
|
Il1f8
|
interleukin 1 family, member 8 |
| chr16_-_58749007 | 0.01 |
ENSMUST00000075361.5
ENSMUST00000205668.2 ENSMUST00000205986.4 |
Olfr181
|
olfactory receptor 181 |
| chr1_-_126758369 | 0.01 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr7_+_140147799 | 0.01 |
ENSMUST00000210973.3
|
Olfr538
|
olfactory receptor 538 |
| chr4_+_45848816 | 0.01 |
ENSMUST00000107782.8
|
Stra6l
|
STRA6-like |
| chr18_+_37568647 | 0.01 |
ENSMUST00000055495.6
|
Pcdhb12
|
protocadherin beta 12 |
| chr2_+_85835884 | 0.01 |
ENSMUST00000111589.3
|
Olfr1032
|
olfactory receptor 1032 |
| chr16_-_44153498 | 0.01 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr16_+_48877762 | 0.01 |
ENSMUST00000168680.2
|
Myh15
|
myosin, heavy chain 15 |
| chr1_+_150195158 | 0.01 |
ENSMUST00000165062.8
ENSMUST00000191228.7 ENSMUST00000186572.7 ENSMUST00000185698.2 |
Pdc
|
phosducin |
| chr17_-_38442081 | 0.01 |
ENSMUST00000087128.2
|
Olfr132
|
olfactory receptor 132 |
| chr19_+_58717319 | 0.01 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr19_-_14575395 | 0.01 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr1_-_185061525 | 0.01 |
ENSMUST00000027921.11
ENSMUST00000110975.8 ENSMUST00000110974.4 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr3_+_69129745 | 0.01 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr7_-_104856692 | 0.01 |
ENSMUST00000216143.2
|
Olfr686
|
olfactory receptor 686 |
| chr19_-_13827773 | 0.01 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr6_-_66656668 | 0.01 |
ENSMUST00000071414.2
|
Vmn1r35
|
vomeronasal 1 receptor 35 |
| chr18_+_37488174 | 0.01 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr19_-_58442866 | 0.01 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr7_-_10488291 | 0.01 |
ENSMUST00000226874.2
ENSMUST00000227003.2 ENSMUST00000228561.2 ENSMUST00000228248.2 ENSMUST00000228526.2 ENSMUST00000228098.2 ENSMUST00000227940.2 ENSMUST00000228374.2 ENSMUST00000227702.2 |
Vmn1r71
|
vomeronasal 1 receptor 71 |
| chr7_-_102822842 | 0.01 |
ENSMUST00000074272.2
|
Olfr591
|
olfactory receptor 591 |
| chr4_-_147787010 | 0.01 |
ENSMUST00000117638.2
|
Zfp534
|
zinc finger protein 534 |
| chr7_-_102805563 | 0.01 |
ENSMUST00000218483.2
|
Olfr589
|
olfactory receptor 589 |
| chr19_+_13316226 | 0.01 |
ENSMUST00000207124.3
|
Olfr1466
|
olfactory receptor 1466 |
| chr9_-_38614969 | 0.01 |
ENSMUST00000215612.2
|
Olfr919
|
olfactory receptor 919 |
| chr6_-_84565613 | 0.01 |
ENSMUST00000204146.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr9_+_38755745 | 0.01 |
ENSMUST00000217350.2
|
Olfr924
|
olfactory receptor 924 |
| chr4_+_90107057 | 0.01 |
ENSMUST00000107129.2
|
Zfp352
|
zinc finger protein 352 |
| chr17_-_43003135 | 0.01 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr4_-_43823866 | 0.01 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr11_-_101857831 | 0.01 |
ENSMUST00000001534.7
|
Sost
|
sclerostin |
| chr4_-_96673423 | 0.01 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr7_+_107413787 | 0.01 |
ENSMUST00000084756.2
|
Olfr467
|
olfactory receptor 467 |
| chr1_-_163552693 | 0.01 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr7_+_10180716 | 0.01 |
ENSMUST00000055964.8
|
Vmn1r67
|
vomeronasal 1 receptor 67 |
| chr9_-_119548098 | 0.01 |
ENSMUST00000084787.6
|
Scn10a
|
sodium channel, voltage-gated, type X, alpha |
| chr7_+_123061497 | 0.01 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr2_-_111452417 | 0.01 |
ENSMUST00000099612.2
|
Olfr1297
|
olfactory receptor 1297 |
| chr7_+_123061535 | 0.01 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr5_+_20112704 | 0.01 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr18_+_37554471 | 0.01 |
ENSMUST00000053073.6
|
Pcdhb11
|
protocadherin beta 11 |
| chr4_+_151012375 | 0.01 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chrX_+_99019176 | 0.01 |
ENSMUST00000113781.8
ENSMUST00000113783.8 ENSMUST00000113779.8 ENSMUST00000113776.8 ENSMUST00000113775.8 ENSMUST00000113780.8 ENSMUST00000113778.8 ENSMUST00000113777.8 ENSMUST00000071453.3 |
Eda
|
ectodysplasin-A |
| chr5_-_131336914 | 0.01 |
ENSMUST00000160609.2
|
Galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.0 | 0.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.0 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |