Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nr0b1
Z-value: 0.75
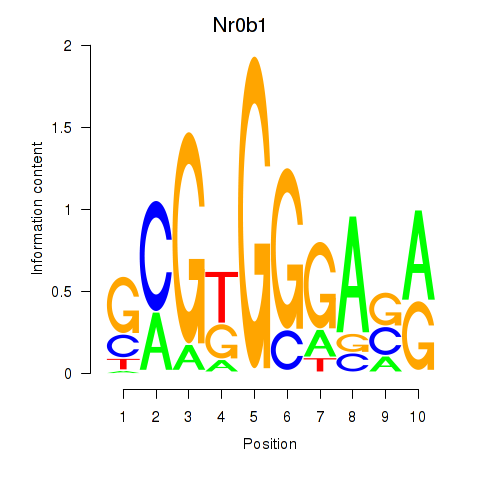
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSMUSG00000025056.5 | nuclear receptor subfamily 0, group B, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm39_v1_chrX_+_85235370_85235388 | 0.60 | 2.8e-01 | Click! |
Activity profile of Nr0b1 motif
Sorted Z-values of Nr0b1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_31070739 | 0.61 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr5_-_107873883 | 0.52 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr2_+_157120946 | 0.50 |
ENSMUST00000116380.9
ENSMUST00000029171.6 |
Rpn2
|
ribophorin II |
| chr19_-_23425757 | 0.32 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr9_-_119812042 | 0.30 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr5_-_31350352 | 0.27 |
ENSMUST00000202758.4
ENSMUST00000114603.8 |
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chrX_-_144288071 | 0.26 |
ENSMUST00000112835.8
ENSMUST00000143610.3 |
Amot
|
angiomotin |
| chr5_-_137530214 | 0.26 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr5_+_31350607 | 0.22 |
ENSMUST00000201535.4
|
Snx17
|
sorting nexin 17 |
| chr1_+_88015524 | 0.20 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr16_-_20440005 | 0.19 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr4_-_123644091 | 0.18 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr8_+_56393488 | 0.18 |
ENSMUST00000000275.10
|
Glra3
|
glycine receptor, alpha 3 subunit |
| chrX_+_13147209 | 0.17 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr4_+_148215339 | 0.17 |
ENSMUST00000084129.9
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr8_-_106434565 | 0.16 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr4_+_116078830 | 0.15 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr8_-_85807281 | 0.15 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr5_+_3393893 | 0.12 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr8_-_85807308 | 0.11 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr10_-_127504416 | 0.11 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr5_-_31350449 | 0.11 |
ENSMUST00000166769.8
|
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr15_-_85918378 | 0.10 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr4_+_53826013 | 0.10 |
ENSMUST00000030127.13
|
Tmem38b
|
transmembrane protein 38B |
| chr19_-_6178171 | 0.10 |
ENSMUST00000154601.8
ENSMUST00000138931.3 |
Snx15
|
sorting nexin 15 |
| chr2_+_27567213 | 0.10 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr19_-_46028060 | 0.09 |
ENSMUST00000056931.14
|
Ldb1
|
LIM domain binding 1 |
| chr6_+_83771985 | 0.08 |
ENSMUST00000113851.8
|
Nagk
|
N-acetylglucosamine kinase |
| chr6_+_120643323 | 0.08 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr7_+_45434755 | 0.07 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr5_-_108134869 | 0.07 |
ENSMUST00000145239.2
ENSMUST00000031198.11 |
Dipk1a
|
divergent protein kinase domain 1A |
| chr6_+_134012916 | 0.07 |
ENSMUST00000164648.2
|
Etv6
|
ets variant 6 |
| chr8_-_71834543 | 0.07 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr5_+_31350566 | 0.07 |
ENSMUST00000031029.15
ENSMUST00000201679.4 |
Snx17
|
sorting nexin 17 |
| chr16_+_56942050 | 0.07 |
ENSMUST00000166897.3
|
Tomm70a
|
translocase of outer mitochondrial membrane 70A |
| chr10_-_128425519 | 0.06 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr4_-_129590609 | 0.06 |
ENSMUST00000102588.10
|
Tmem39b
|
transmembrane protein 39b |
| chr12_+_3857077 | 0.05 |
ENSMUST00000174817.8
|
Dnmt3a
|
DNA methyltransferase 3A |
| chr15_+_98532624 | 0.05 |
ENSMUST00000003442.9
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chrX_-_47123719 | 0.05 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr15_-_81931783 | 0.04 |
ENSMUST00000080622.9
|
Snu13
|
SNU13 homolog, small nuclear ribonucleoprotein (U4/U6.U5) |
| chr11_-_88608958 | 0.04 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr14_-_61677258 | 0.04 |
ENSMUST00000022496.9
|
Kpna3
|
karyopherin (importin) alpha 3 |
| chr11_-_115503704 | 0.04 |
ENSMUST00000106506.8
|
Mif4gd
|
MIF4G domain containing |
| chr19_-_6127211 | 0.04 |
ENSMUST00000160590.2
ENSMUST00000025711.13 |
Vps51
|
VPS51 GARP complex subunit |
| chr11_-_115503316 | 0.04 |
ENSMUST00000106507.9
|
Mif4gd
|
MIF4G domain containing |
| chr4_+_116078874 | 0.04 |
ENSMUST00000106490.3
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr11_+_105072619 | 0.03 |
ENSMUST00000092537.10
ENSMUST00000015107.13 ENSMUST00000145048.8 |
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr7_-_80020590 | 0.03 |
ENSMUST00000206212.2
|
Man2a2
|
mannosidase 2, alpha 2 |
| chr18_+_35963353 | 0.03 |
ENSMUST00000235169.2
|
Cxxc5
|
CXXC finger 5 |
| chr9_-_108888779 | 0.03 |
ENSMUST00000061973.5
|
Trex1
|
three prime repair exonuclease 1 |
| chr5_-_99185201 | 0.03 |
ENSMUST00000161490.8
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr11_-_120238917 | 0.03 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr19_+_37423198 | 0.03 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr3_+_105359641 | 0.03 |
ENSMUST00000098761.10
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3 |
| chr6_+_85164420 | 0.03 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr10_+_36383008 | 0.02 |
ENSMUST00000168572.8
|
Hs3st5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr10_-_81186222 | 0.02 |
ENSMUST00000020454.11
ENSMUST00000105324.9 ENSMUST00000154609.3 ENSMUST00000105323.8 |
Hmg20b
|
high mobility group 20B |
| chr17_-_25652750 | 0.02 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr2_-_131194754 | 0.02 |
ENSMUST00000059372.11
|
Rnf24
|
ring finger protein 24 |
| chr4_-_102883905 | 0.02 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr10_-_81186137 | 0.02 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr8_-_71308229 | 0.02 |
ENSMUST00000212086.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr17_+_88837540 | 0.02 |
ENSMUST00000038551.8
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr11_-_69871320 | 0.02 |
ENSMUST00000143175.2
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr2_-_144174066 | 0.02 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr11_+_73819052 | 0.02 |
ENSMUST00000205791.2
|
Olfr396-ps1
|
olfactory receptor 396, pseudogene 1 |
| chr17_+_23898223 | 0.02 |
ENSMUST00000024699.4
ENSMUST00000232719.2 |
Cldn6
|
claudin 6 |
| chr1_+_140173787 | 0.02 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr8_-_71308040 | 0.01 |
ENSMUST00000212509.3
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr6_-_72212547 | 0.01 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr5_-_99184894 | 0.01 |
ENSMUST00000031277.7
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr19_-_46027881 | 0.01 |
ENSMUST00000137771.2
|
Ldb1
|
LIM domain binding 1 |
| chr2_+_153684901 | 0.01 |
ENSMUST00000175856.3
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr15_+_98532866 | 0.01 |
ENSMUST00000230490.2
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr2_-_152256947 | 0.01 |
ENSMUST00000099207.5
|
Zcchc3
|
zinc finger, CCHC domain containing 3 |
| chr4_+_116078787 | 0.01 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr2_+_4722956 | 0.01 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr2_+_81883566 | 0.01 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr2_-_157121440 | 0.01 |
ENSMUST00000143663.2
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr2_-_144173615 | 0.01 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr11_+_19874403 | 0.01 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr3_-_86827640 | 0.01 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chrX_-_37653396 | 0.00 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr5_+_30972067 | 0.00 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr9_-_110571645 | 0.00 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr8_+_106434901 | 0.00 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr11_-_52165682 | 0.00 |
ENSMUST00000238914.2
|
Tcf7
|
transcription factor 7, T cell specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr0b1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |