Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nr2c1
Z-value: 0.32
Transcription factors associated with Nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c1
|
ENSMUSG00000005897.15 | nuclear receptor subfamily 2, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c1 | mm39_v1_chr10_+_93983844_93983885 | -0.73 | 1.6e-01 | Click! |
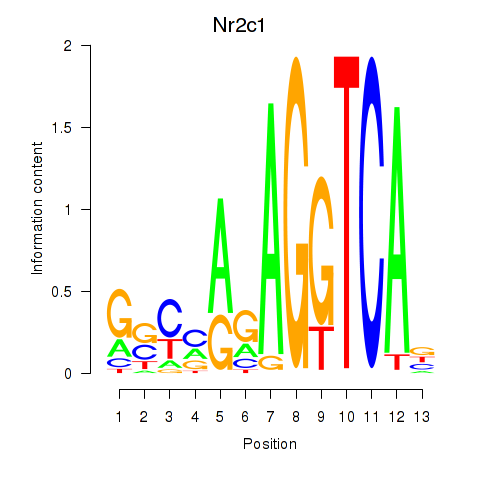
Activity profile of Nr2c1 motif
Sorted Z-values of Nr2c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_73476629 | 0.29 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr6_-_83030759 | 0.25 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr8_+_121395047 | 0.22 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr17_-_26014613 | 0.21 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chr10_-_119075910 | 0.19 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr17_-_27816151 | 0.16 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr5_+_145060489 | 0.16 |
ENSMUST00000136074.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr14_+_51162425 | 0.16 |
ENSMUST00000049411.12
ENSMUST00000136753.8 ENSMUST00000154288.3 |
Apex1
|
apurinic/apyrimidinic endonuclease 1 |
| chr6_-_83031358 | 0.16 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr14_-_52252003 | 0.14 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chr16_-_4340920 | 0.14 |
ENSMUST00000090500.10
ENSMUST00000023161.8 |
Srl
|
sarcalumenin |
| chr9_-_60594742 | 0.14 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_-_120047868 | 0.13 |
ENSMUST00000112648.8
|
Dbi
|
diazepam binding inhibitor |
| chr5_-_110928436 | 0.13 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr9_-_50571080 | 0.12 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr5_-_110927803 | 0.12 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr19_-_6899173 | 0.12 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr3_+_115681486 | 0.12 |
ENSMUST00000189799.7
ENSMUST00000200258.2 |
Dph5
|
diphthamide biosynthesis 5 |
| chr4_+_118286898 | 0.11 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr3_+_103716836 | 0.11 |
ENSMUST00000076599.8
ENSMUST00000106824.8 ENSMUST00000106823.8 ENSMUST00000047285.7 |
Ap4b1
|
adaptor-related protein complex AP-4, beta 1 |
| chr14_+_122771734 | 0.11 |
ENSMUST00000154206.8
ENSMUST00000038374.13 ENSMUST00000135578.8 |
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide |
| chr19_-_6899121 | 0.10 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr7_+_101043568 | 0.10 |
ENSMUST00000098243.4
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_46343291 | 0.10 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr15_-_102579463 | 0.09 |
ENSMUST00000185641.7
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) |
| chr2_+_92205651 | 0.09 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr5_+_137628377 | 0.09 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr3_+_95226093 | 0.09 |
ENSMUST00000139866.2
|
Cers2
|
ceramide synthase 2 |
| chr11_-_62172164 | 0.09 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr11_+_116744578 | 0.09 |
ENSMUST00000021173.14
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr4_+_97665843 | 0.08 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr17_+_35413415 | 0.08 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr1_-_164285914 | 0.08 |
ENSMUST00000027863.13
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chrX_-_101232978 | 0.08 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr17_-_13211374 | 0.07 |
ENSMUST00000159551.8
ENSMUST00000160781.8 |
Wtap
|
Wilms tumour 1-associating protein |
| chr3_+_32583602 | 0.07 |
ENSMUST00000091257.11
|
Mfn1
|
mitofusin 1 |
| chr2_-_30720345 | 0.07 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr19_+_5012362 | 0.07 |
ENSMUST00000236917.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr15_-_79025387 | 0.07 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr6_-_119365632 | 0.06 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr12_-_87194658 | 0.06 |
ENSMUST00000037788.6
|
Pomt2
|
protein-O-mannosyltransferase 2 |
| chr11_-_107228382 | 0.06 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr2_+_22785534 | 0.06 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr5_+_135216090 | 0.06 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr10_+_58207229 | 0.06 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_+_25774070 | 0.06 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr14_-_51162083 | 0.06 |
ENSMUST00000160375.8
ENSMUST00000162177.8 |
Osgep
|
O-sialoglycoprotein endopeptidase |
| chr19_+_5012396 | 0.06 |
ENSMUST00000235229.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr15_-_76396151 | 0.06 |
ENSMUST00000023214.11
|
Dgat1
|
diacylglycerol O-acyltransferase 1 |
| chr3_+_68479578 | 0.06 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr14_-_52252432 | 0.05 |
ENSMUST00000226527.2
|
Zfp219
|
zinc finger protein 219 |
| chr1_-_58625431 | 0.05 |
ENSMUST00000161000.2
ENSMUST00000161600.8 |
Fam126b
|
family with sequence similarity 126, member B |
| chr2_-_164646794 | 0.05 |
ENSMUST00000103094.11
ENSMUST00000017451.7 |
Acot8
|
acyl-CoA thioesterase 8 |
| chr4_-_11076159 | 0.05 |
ENSMUST00000058183.9
|
Ndufaf6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr13_+_104424359 | 0.05 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr3_-_79535966 | 0.05 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr1_-_120048667 | 0.05 |
ENSMUST00000151708.3
|
Dbi
|
diazepam binding inhibitor |
| chr1_-_74583434 | 0.04 |
ENSMUST00000189257.7
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr7_+_79836581 | 0.04 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr11_-_76468527 | 0.04 |
ENSMUST00000176179.8
|
Abr
|
active BCR-related gene |
| chr2_+_109522781 | 0.04 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr4_+_150938376 | 0.04 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr16_-_11072025 | 0.04 |
ENSMUST00000080030.14
|
Gspt1
|
G1 to S phase transition 1 |
| chr3_+_32583681 | 0.04 |
ENSMUST00000147350.8
|
Mfn1
|
mitofusin 1 |
| chr2_+_22785593 | 0.04 |
ENSMUST00000152170.8
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr7_-_126483851 | 0.04 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr1_+_180553569 | 0.04 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr4_-_130068484 | 0.04 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr1_-_74323795 | 0.04 |
ENSMUST00000178235.8
ENSMUST00000006462.14 |
Aamp
|
angio-associated migratory protein |
| chr11_+_75084609 | 0.04 |
ENSMUST00000102514.4
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chr16_+_20367327 | 0.04 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr17_+_35069347 | 0.03 |
ENSMUST00000097343.11
ENSMUST00000173357.8 ENSMUST00000173065.8 ENSMUST00000165953.3 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr9_-_106035308 | 0.03 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr14_-_52252318 | 0.03 |
ENSMUST00000228051.2
|
Zfp219
|
zinc finger protein 219 |
| chr16_-_32306683 | 0.03 |
ENSMUST00000042042.9
|
Slc51a
|
solute carrier family 51, alpha subunit |
| chr3_-_8988854 | 0.03 |
ENSMUST00000042148.6
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr15_+_79975520 | 0.03 |
ENSMUST00000009728.13
ENSMUST00000009727.12 |
Syngr1
|
synaptogyrin 1 |
| chr7_-_28597523 | 0.03 |
ENSMUST00000216863.2
|
Actn4
|
actinin alpha 4 |
| chr14_+_58036093 | 0.03 |
ENSMUST00000111267.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr5_-_92231314 | 0.03 |
ENSMUST00000169094.8
ENSMUST00000167918.8 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr4_-_82777746 | 0.03 |
ENSMUST00000156055.2
ENSMUST00000030110.15 |
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr12_+_87194476 | 0.03 |
ENSMUST00000063117.10
ENSMUST00000220574.2 |
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr8_-_85663976 | 0.03 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr16_-_11072131 | 0.03 |
ENSMUST00000167571.8
|
Gspt1
|
G1 to S phase transition 1 |
| chr12_+_87193922 | 0.03 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr8_+_96551974 | 0.03 |
ENSMUST00000074053.6
|
Sap18b
|
Sin3-associated polypeptide 18B |
| chr3_-_107538996 | 0.03 |
ENSMUST00000064759.7
|
Strip1
|
striatin interacting protein 1 |
| chr7_-_45570674 | 0.03 |
ENSMUST00000210939.2
|
Emp3
|
epithelial membrane protein 3 |
| chr17_-_35454729 | 0.03 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr10_+_41395410 | 0.02 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr3_-_58433313 | 0.02 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr13_+_55468313 | 0.02 |
ENSMUST00000021942.8
|
Prelid1
|
PRELI domain containing 1 |
| chr11_+_4936824 | 0.02 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr19_+_5012336 | 0.02 |
ENSMUST00000237974.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr7_+_45546365 | 0.02 |
ENSMUST00000069772.16
ENSMUST00000210503.2 ENSMUST00000209350.2 |
Tmem143
|
transmembrane protein 143 |
| chr15_+_86098660 | 0.02 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr1_+_74545203 | 0.02 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr4_+_156300325 | 0.02 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr1_+_58625539 | 0.02 |
ENSMUST00000027193.9
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr17_-_13211075 | 0.02 |
ENSMUST00000159986.8
ENSMUST00000007007.14 |
Wtap
|
Wilms tumour 1-associating protein |
| chr17_-_34247016 | 0.02 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr4_+_117109204 | 0.02 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr2_+_158636727 | 0.01 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr7_-_45570254 | 0.01 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr15_+_81695615 | 0.01 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr16_+_41353360 | 0.01 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr8_-_124621483 | 0.01 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr17_+_38110779 | 0.01 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr1_+_163942833 | 0.01 |
ENSMUST00000162746.2
|
Selp
|
selectin, platelet |
| chr1_-_74323536 | 0.01 |
ENSMUST00000190008.2
|
Aamp
|
angio-associated migratory protein |
| chr1_-_58625350 | 0.01 |
ENSMUST00000038372.14
ENSMUST00000097724.10 |
Fam126b
|
family with sequence similarity 126, member B |
| chr12_-_115083839 | 0.01 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr11_+_83741657 | 0.01 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr8_+_55024446 | 0.01 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr4_-_132459762 | 0.01 |
ENSMUST00000045550.5
|
Xkr8
|
X-linked Kx blood group related 8 |
| chr11_+_83741689 | 0.01 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr11_-_50216426 | 0.01 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr2_-_128890092 | 0.01 |
ENSMUST00000145798.4
|
Vinac1
|
vinculin/alpha-catenin family member 1 |
| chr13_-_55718899 | 0.01 |
ENSMUST00000225240.3
ENSMUST00000021957.8 |
Fam193b
|
family with sequence similarity 193, member B |
| chr2_+_110427643 | 0.01 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr14_+_58036071 | 0.01 |
ENSMUST00000111268.8
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr7_-_45570538 | 0.01 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr16_-_58850641 | 0.01 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chr13_-_73476561 | 0.01 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr3_+_129007599 | 0.01 |
ENSMUST00000042587.12
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr4_+_117109148 | 0.01 |
ENSMUST00000062824.12
|
Tmem53
|
transmembrane protein 53 |
| chr11_-_99494134 | 0.00 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr8_-_123187406 | 0.00 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr11_-_100098333 | 0.00 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr10_-_129010923 | 0.00 |
ENSMUST00000169800.4
|
Olfr772
|
olfactory receptor 772 |
| chr3_+_58433236 | 0.00 |
ENSMUST00000029387.15
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr3_+_129007917 | 0.00 |
ENSMUST00000174623.3
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr11_+_98274637 | 0.00 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chrX_-_58612709 | 0.00 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr9_-_65298934 | 0.00 |
ENSMUST00000068307.4
|
Kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr19_+_13422378 | 0.00 |
ENSMUST00000096203.2
|
Olfr1471
|
olfactory receptor 1471 |
| chr4_-_41517326 | 0.00 |
ENSMUST00000030152.13
ENSMUST00000095126.5 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr9_-_40012784 | 0.00 |
ENSMUST00000056795.4
|
Olfr984
|
olfactory receptor 984 |
| chr17_+_75772567 | 0.00 |
ENSMUST00000234660.2
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr15_-_102274781 | 0.00 |
ENSMUST00000078508.7
|
Sp7
|
Sp7 transcription factor 7 |
| chr10_+_4382467 | 0.00 |
ENSMUST00000095893.11
ENSMUST00000118544.8 ENSMUST00000117489.8 |
Armt1
|
acidic residue methyltransferase 1 |
| chr3_+_86131970 | 0.00 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr9_+_26645024 | 0.00 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr1_+_166081755 | 0.00 |
ENSMUST00000194964.6
ENSMUST00000192638.6 ENSMUST00000192426.6 ENSMUST00000195557.6 ENSMUST00000192732.6 ENSMUST00000193860.2 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr2_-_62404195 | 0.00 |
ENSMUST00000174234.8
ENSMUST00000000402.16 ENSMUST00000174448.8 ENSMUST00000102732.10 |
Fap
|
fibroblast activation protein |
| chr12_+_35975619 | 0.00 |
ENSMUST00000042101.5
ENSMUST00000154042.2 |
Agr3
|
anterior gradient 3 |
| chr4_+_127066667 | 0.00 |
ENSMUST00000106094.9
|
Dlgap3
|
DLG associated protein 3 |
| chr7_+_30193047 | 0.00 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr12_-_83534482 | 0.00 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr9_+_26645141 | 0.00 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr10_+_75152908 | 0.00 |
ENSMUST00000219044.2
|
Adora2a
|
adenosine A2a receptor |
| chr7_+_43474819 | 0.00 |
ENSMUST00000107967.3
|
Klk6
|
kallikrein related-peptidase 6 |
| chr9_-_35755446 | 0.00 |
ENSMUST00000054175.7
|
Pate5
|
prostate and testis expressed 5 |
| chr10_-_14581203 | 0.00 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr6_-_128252540 | 0.00 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr2_+_106525938 | 0.00 |
ENSMUST00000016530.14
|
Mpped2
|
metallophosphoesterase domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin A metabolic process(GO:1901738) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.0 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.0 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |