Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nr2e1
Z-value: 0.18
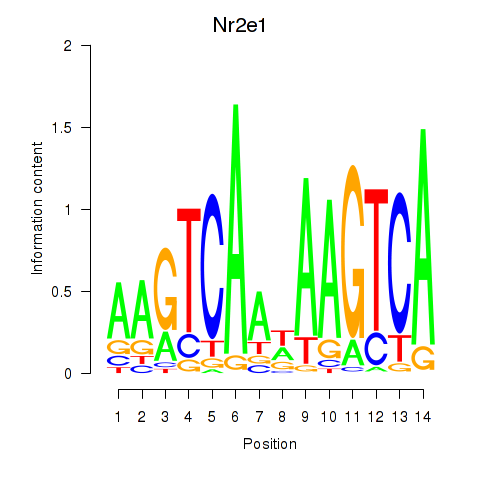
Transcription factors associated with Nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e1
|
ENSMUSG00000019803.12 | nuclear receptor subfamily 2, group E, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e1 | mm39_v1_chr10_-_42459624_42459635 | -0.65 | 2.3e-01 | Click! |
Activity profile of Nr2e1 motif
Sorted Z-values of Nr2e1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_28813125 | 0.07 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr18_+_36926929 | 0.06 |
ENSMUST00000001419.10
|
Zmat2
|
zinc finger, matrin type 2 |
| chrM_+_7006 | 0.05 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr4_+_28813152 | 0.05 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chrX_-_94701983 | 0.05 |
ENSMUST00000119640.8
ENSMUST00000120620.8 ENSMUST00000044382.7 |
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr10_-_61619790 | 0.05 |
ENSMUST00000020283.5
|
Macroh2a2
|
macroH2A.2 histone |
| chr7_-_80055168 | 0.05 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr9_-_71070506 | 0.04 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr11_+_68858942 | 0.04 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr15_-_97603664 | 0.03 |
ENSMUST00000023104.7
|
Rpap3
|
RNA polymerase II associated protein 3 |
| chr3_-_146487102 | 0.03 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr10_+_78412783 | 0.02 |
ENSMUST00000219588.2
|
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chrY_-_9132561 | 0.02 |
ENSMUST00000171947.3
|
Gm21292
|
predicted gene, 21292 |
| chr14_+_28740162 | 0.02 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr2_-_85632888 | 0.02 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr14_+_26414422 | 0.01 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr9_-_32454157 | 0.01 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr10_+_18345706 | 0.01 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr15_-_10485976 | 0.01 |
ENSMUST00000169050.8
ENSMUST00000022855.12 |
Brix1
|
BRX1, biogenesis of ribosomes |
| chr17_+_48037758 | 0.01 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr18_+_34973605 | 0.01 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr6_-_58418303 | 0.01 |
ENSMUST00000228577.2
ENSMUST00000227466.2 |
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr1_-_75196496 | 0.01 |
ENSMUST00000186758.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr4_+_33924632 | 0.01 |
ENSMUST00000057188.7
|
Cnr1
|
cannabinoid receptor 1 (brain) |
| chr6_+_65567373 | 0.01 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr18_-_73836810 | 0.01 |
ENSMUST00000025393.14
|
Smad4
|
SMAD family member 4 |
| chrX_+_48761210 | 0.01 |
ENSMUST00000214375.2
ENSMUST00000214273.2 |
Olfr1320
|
olfactory receptor 1320 |
| chr19_-_11261177 | 0.01 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr2_-_111166421 | 0.01 |
ENSMUST00000099618.2
|
Olfr1282
|
olfactory receptor 1282 |
| chr13_+_24023428 | 0.01 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr8_-_62355690 | 0.01 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr5_+_16996808 | 0.01 |
ENSMUST00000211738.2
|
Gm28710
|
predicted gene 28710 |
| chr4_-_35845204 | 0.01 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr6_-_126916487 | 0.01 |
ENSMUST00000144954.5
ENSMUST00000112220.8 ENSMUST00000112221.8 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr8_+_36956345 | 0.01 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr4_+_135455427 | 0.01 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr4_-_62005498 | 0.01 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr5_-_123185029 | 0.01 |
ENSMUST00000045843.15
|
Morn3
|
MORN repeat containing 3 |
| chr2_-_22930188 | 0.01 |
ENSMUST00000114544.10
ENSMUST00000139038.8 ENSMUST00000126112.8 ENSMUST00000178908.2 ENSMUST00000078977.14 ENSMUST00000140164.8 ENSMUST00000149719.8 |
Abi1
|
abl interactor 1 |
| chr14_-_33737131 | 0.01 |
ENSMUST00000227795.2
|
Gm30083
|
predicted gene, 30083 |
| chr13_+_24023386 | 0.01 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr4_-_59960659 | 0.00 |
ENSMUST00000075973.3
|
Mup4
|
major urinary protein 4 |
| chrY_-_79161056 | 0.00 |
ENSMUST00000179922.2
|
Gm20917
|
predicted gene, 20917 |
| chr6_-_141719536 | 0.00 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr2_+_69050315 | 0.00 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr12_+_86781141 | 0.00 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr14_-_54754810 | 0.00 |
ENSMUST00000023873.12
|
Prmt5
|
protein arginine N-methyltransferase 5 |
| chr18_+_77369654 | 0.00 |
ENSMUST00000096547.11
ENSMUST00000148341.9 ENSMUST00000123410.9 |
Loxhd1
|
lipoxygenase homology domains 1 |
| chr6_+_120070307 | 0.00 |
ENSMUST00000112711.9
|
Ninj2
|
ninjurin 2 |
| chr7_+_114367971 | 0.00 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr12_+_86781154 | 0.00 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr15_+_41694317 | 0.00 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr5_-_123185073 | 0.00 |
ENSMUST00000031437.14
|
Morn3
|
MORN repeat containing 3 |
| chrY_-_85540787 | 0.00 |
ENSMUST00000178889.2
|
Gm20854
|
predicted gene, 20854 |
| chr3_-_24837772 | 0.00 |
ENSMUST00000203414.2
|
Naaladl2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr8_+_71156071 | 0.00 |
ENSMUST00000212436.2
|
Iqcn
|
IQ motif containing N |
| chr10_-_23869379 | 0.00 |
ENSMUST00000078532.3
|
Taar7a
|
trace amine-associated receptor 7A |
| chr3_+_96432479 | 0.00 |
ENSMUST00000049208.11
|
Hjv
|
hemojuvelin BMP co-receptor |
| chrY_+_68550741 | 0.00 |
ENSMUST00000177765.2
|
Gm20816
|
predicted gene, 20816 |
| chrY_-_78847812 | 0.00 |
ENSMUST00000180324.2
|
Gm20806
|
predicted gene, 20806 |
| chr5_-_87288177 | 0.00 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chrX_-_133012457 | 0.00 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chrY_+_21166083 | 0.00 |
ENSMUST00000178234.2
|
Gm20909
|
predicted gene, 20909 |
| chrY_-_53413354 | 0.00 |
ENSMUST00000179137.2
|
Gm20747
|
predicted gene, 20747 |
| chrY_+_24411927 | 0.00 |
ENSMUST00000179663.2
|
Gm20809
|
predicted gene, 20809 |
| chr4_-_62069046 | 0.00 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chrY_-_77972959 | 0.00 |
ENSMUST00000178900.2
|
Gm20867
|
predicted gene, 20867 |
| chrX_+_30080456 | 0.00 |
ENSMUST00000177917.2
|
Gm14632
|
predicted gene 14632 |
| chrX_-_30540953 | 0.00 |
ENSMUST00000177566.2
|
Gm10487
|
predicted gene 10487 |
| chrY_-_84574663 | 0.00 |
ENSMUST00000179523.2
|
Gm21394
|
predicted gene, 21394 |
| chrX_-_28733497 | 0.00 |
ENSMUST00000178672.2
|
Gm10096
|
predicted gene 10096 |
| chrX_-_27725713 | 0.00 |
ENSMUST00000178152.2
|
Gm10058
|
predicted gene 10058 |
| chr14_+_33775423 | 0.00 |
ENSMUST00000058725.5
|
Antxrl
|
anthrax toxin receptor-like |
| chrY_+_21242966 | 0.00 |
ENSMUST00000179095.2
|
Gm20865
|
predicted gene, 20865 |
| chr4_-_61437704 | 0.00 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chrX_+_34669447 | 0.00 |
ENSMUST00000169396.3
|
Gm14819
|
predicted gene 14819 |
| chrX_-_27378797 | 0.00 |
ENSMUST00000178745.2
|
Gm10230
|
predicted gene 10230 |
| chrX_-_28386687 | 0.00 |
ENSMUST00000178966.2
|
Gm10147
|
predicted gene 10147 |
| chrX_-_26004758 | 0.00 |
ENSMUST00000168002.3
|
Gm5168
|
predicted gene 5168 |
| chrY_-_66751879 | 0.00 |
ENSMUST00000178761.2
|
Gm20852
|
predicted gene, 20852 |
| chr6_+_126916919 | 0.00 |
ENSMUST00000032497.7
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chrX_-_24639814 | 0.00 |
ENSMUST00000178986.2
|
Gm5935
|
predicted gene 5935 |
| chrY_-_65214536 | 0.00 |
ENSMUST00000177663.2
|
Gm20924
|
predicted gene, 20924 |
| chrY_-_70283393 | 0.00 |
ENSMUST00000180179.2
|
Gm21118
|
predicted gene, 21118 |
| chrY_-_9345654 | 0.00 |
ENSMUST00000179590.2
|
Gm21812
|
predicted gene, 21812 |
| chr11_+_49039086 | 0.00 |
ENSMUST00000059379.2
|
Olfr1395
|
olfactory receptor 1395 |
| chr4_-_60697274 | 0.00 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr2_+_14828903 | 0.00 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_-_60457902 | 0.00 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr3_+_130411294 | 0.00 |
ENSMUST00000163620.8
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr5_-_87054796 | 0.00 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr4_-_60777462 | 0.00 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr2_-_22930104 | 0.00 |
ENSMUST00000153931.8
ENSMUST00000123948.8 |
Abi1
|
abl interactor 1 |
| chr7_+_131144596 | 0.00 |
ENSMUST00000046093.6
|
Hmx3
|
H6 homeobox 3 |
| chrY_-_7636973 | 0.00 |
ENSMUST00000179256.2
|
Gm20826
|
predicted gene, 20826 |
| chr19_+_13387935 | 0.00 |
ENSMUST00000077538.2
|
Olfr1469
|
olfactory receptor 1469 |
| chr4_-_119272640 | 0.00 |
ENSMUST00000238293.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chrY_+_9783380 | 0.00 |
ENSMUST00000180352.2
|
Gm20821
|
predicted gene, 20821 |
| chrY_-_9200643 | 0.00 |
ENSMUST00000179595.2
|
Gm21721
|
predicted gene, 21721 |
| chr17_+_32904629 | 0.00 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr2_-_90083330 | 0.00 |
ENSMUST00000099752.2
|
Olfr142
|
olfactory receptor 142 |
| chr9_+_8544228 | 0.00 |
ENSMUST00000214596.2
|
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chrY_+_12383184 | 0.00 |
ENSMUST00000179220.2
|
Gm20812
|
predicted gene, 20812 |
| chrX_+_48552803 | 0.00 |
ENSMUST00000130558.8
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr17_-_20403359 | 0.00 |
ENSMUST00000095636.2
|
Fpr-rs6
|
formyl peptide receptor, related sequence 6 |
| chrY_-_10323434 | 0.00 |
ENSMUST00000179558.2
|
Gm20834
|
predicted gene, 20834 |
| chrY_-_6917117 | 0.00 |
ENSMUST00000178016.2
|
Gm20830
|
predicted gene, 20830 |
| chr4_+_60003438 | 0.00 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chr2_-_22930149 | 0.00 |
ENSMUST00000091394.13
ENSMUST00000093171.13 |
Abi1
|
abl interactor 1 |
| chrY_-_7167777 | 0.00 |
ENSMUST00000178447.2
|
Gm21244
|
predicted gene, 21244 |
| chrY_-_15883245 | 0.00 |
ENSMUST00000165756.2
|
Gm20822
|
predicted gene, 20822 |
| chr4_-_61259801 | 0.00 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr17_+_32904601 | 0.00 |
ENSMUST00000168171.8
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr4_-_61259997 | 0.00 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chrY_-_11197848 | 0.00 |
ENSMUST00000170889.2
|
Gm20828
|
predicted gene, 20828 |
| chr4_-_96029375 | 0.00 |
ENSMUST00000097972.5
|
Cyp2j12
|
cytochrome P450, family 2, subfamily j, polypeptide 12 |
| chr5_-_72738597 | 0.00 |
ENSMUST00000073528.4
|
Zar1
|
zygote arrest 1 |
| chr14_+_31750946 | 0.00 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.0 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008046 | GPI-linked ephrin receptor activity(GO:0005004) axon guidance receptor activity(GO:0008046) |