Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nr2e3
Z-value: 1.65
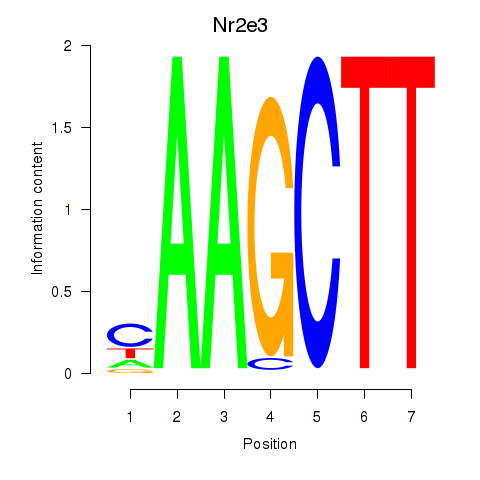
Transcription factors associated with Nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e3
|
ENSMUSG00000032292.9 | nuclear receptor subfamily 2, group E, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e3 | mm39_v1_chr9_-_59857355_59857407 | -0.32 | 6.0e-01 | Click! |
Activity profile of Nr2e3 motif
Sorted Z-values of Nr2e3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_35347983 | 1.76 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr13_-_29137673 | 1.21 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr7_+_100143250 | 1.11 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr3_-_90603013 | 1.09 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr9_+_113641615 | 0.93 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chrX_+_168662592 | 0.92 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chrX_+_41241049 | 0.88 |
ENSMUST00000128799.3
|
Stag2
|
stromal antigen 2 |
| chr2_-_164197987 | 0.88 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr5_-_9047324 | 0.84 |
ENSMUST00000003720.5
|
Crot
|
carnitine O-octanoyltransferase |
| chr7_-_101714251 | 0.75 |
ENSMUST00000130074.2
ENSMUST00000131104.3 ENSMUST00000096639.12 |
Rnf121
|
ring finger protein 121 |
| chr7_+_101714943 | 0.72 |
ENSMUST00000094130.4
ENSMUST00000084843.10 |
Xndc1
Xntrpc
|
Xrcc1 N-terminal domain containing 1 Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
| chr4_+_102617495 | 0.70 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr3_-_88410495 | 0.69 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr17_+_87270504 | 0.67 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr4_-_154721288 | 0.67 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr4_+_123176570 | 0.67 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr7_-_110673269 | 0.66 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr8_-_58106027 | 0.63 |
ENSMUST00000110316.3
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr8_-_58106057 | 0.62 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr4_+_102617332 | 0.62 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr19_+_8748347 | 0.61 |
ENSMUST00000010248.4
|
Tmem223
|
transmembrane protein 223 |
| chrX_+_37689503 | 0.56 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr2_-_164198427 | 0.55 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr4_-_135699205 | 0.55 |
ENSMUST00000105852.8
|
Lypla2
|
lysophospholipase 2 |
| chr16_+_22926504 | 0.53 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr4_-_136776006 | 0.50 |
ENSMUST00000049583.8
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chr4_+_11579648 | 0.50 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr2_-_90735171 | 0.50 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr7_+_101714692 | 0.50 |
ENSMUST00000106950.8
ENSMUST00000146450.8 |
Xndc1
|
Xrcc1 N-terminal domain containing 1 |
| chr6_-_129599645 | 0.49 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr2_-_101479846 | 0.48 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr7_+_120634834 | 0.48 |
ENSMUST00000207351.2
|
Mettl9
|
methyltransferase like 9 |
| chr8_+_89015705 | 0.48 |
ENSMUST00000171456.9
|
Adcy7
|
adenylate cyclase 7 |
| chr1_-_165762469 | 0.47 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr12_-_110669076 | 0.47 |
ENSMUST00000155242.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_34578842 | 0.47 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr15_+_6329263 | 0.47 |
ENSMUST00000078019.13
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr16_-_56984137 | 0.46 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
| chr3_-_10273628 | 0.45 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr16_-_22258469 | 0.44 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr2_+_90735077 | 0.44 |
ENSMUST00000111464.8
ENSMUST00000090682.4 |
Kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr8_-_61407760 | 0.44 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr1_-_37575313 | 0.44 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr7_-_19005721 | 0.43 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr11_-_100986192 | 0.41 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr1_+_177273226 | 0.41 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr4_-_129534403 | 0.40 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr13_+_21663077 | 0.39 |
ENSMUST00000062609.6
ENSMUST00000225845.2 |
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr15_-_81756076 | 0.37 |
ENSMUST00000023117.10
|
Phf5a
|
PHD finger protein 5A |
| chr9_-_111086528 | 0.35 |
ENSMUST00000199404.2
|
Mlh1
|
mutL homolog 1 |
| chr4_-_49521036 | 0.35 |
ENSMUST00000057829.4
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr9_+_106080307 | 0.35 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr3_-_143908060 | 0.35 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr18_+_48179711 | 0.34 |
ENSMUST00000235307.2
|
Eno1b
|
enolase 1B, retrotransposed |
| chr16_+_31241085 | 0.33 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr2_+_24839758 | 0.33 |
ENSMUST00000028350.9
|
Zmynd19
|
zinc finger, MYND domain containing 19 |
| chr4_+_119052693 | 0.33 |
ENSMUST00000097908.4
|
Svbp
|
small vasohibin binding protein |
| chr15_+_81756671 | 0.32 |
ENSMUST00000135198.2
ENSMUST00000157003.8 ENSMUST00000229068.2 |
Aco2
|
aconitase 2, mitochondrial |
| chr14_-_78866714 | 0.31 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr5_+_76331727 | 0.31 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165 |
| chr2_+_83554741 | 0.31 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr3_-_143908111 | 0.29 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr12_-_54842488 | 0.29 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chr9_+_65494469 | 0.29 |
ENSMUST00000239405.2
ENSMUST00000047099.13 ENSMUST00000131483.3 ENSMUST00000141046.3 |
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chr11_+_58062467 | 0.28 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr4_-_25281750 | 0.28 |
ENSMUST00000038705.8
ENSMUST00000102994.10 |
Ufl1
|
UFM1 specific ligase 1 |
| chr1_-_40829801 | 0.27 |
ENSMUST00000039672.6
|
Mfsd9
|
major facilitator superfamily domain containing 9 |
| chr11_-_55075855 | 0.27 |
ENSMUST00000039305.6
|
Slc36a2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr13_+_25127127 | 0.26 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr17_+_87270707 | 0.26 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr11_-_80030735 | 0.25 |
ENSMUST00000136996.2
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr11_+_93934940 | 0.25 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr4_+_119052548 | 0.25 |
ENSMUST00000106345.3
|
Svbp
|
small vasohibin binding protein |
| chrX_+_7656251 | 0.24 |
ENSMUST00000140540.2
|
Gripap1
|
GRIP1 associated protein 1 |
| chr7_-_118091135 | 0.23 |
ENSMUST00000178344.3
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
| chr11_+_19874403 | 0.23 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr9_-_20085353 | 0.21 |
ENSMUST00000215984.3
|
Olfr870
|
olfactory receptor 870 |
| chr2_-_26917921 | 0.21 |
ENSMUST00000102890.11
ENSMUST00000153388.2 ENSMUST00000045702.6 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr5_-_22755274 | 0.20 |
ENSMUST00000030872.12
|
Orc5
|
origin recognition complex, subunit 5 |
| chr12_+_86999366 | 0.20 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr6_+_140369763 | 0.20 |
ENSMUST00000087622.6
|
Plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr8_+_84335176 | 0.20 |
ENSMUST00000212300.2
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr15_+_6329278 | 0.18 |
ENSMUST00000159046.2
ENSMUST00000161040.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr9_+_65494429 | 0.17 |
ENSMUST00000134538.9
|
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chr7_+_49408847 | 0.17 |
ENSMUST00000085272.7
ENSMUST00000207895.2 |
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chrX_-_133012600 | 0.17 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr5_-_104169785 | 0.16 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr11_+_19874354 | 0.15 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr1_-_97904958 | 0.15 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr18_-_66155651 | 0.14 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr4_-_109522502 | 0.13 |
ENSMUST00000063531.5
|
Cdkn2c
|
cyclin dependent kinase inhibitor 2C |
| chr6_-_13839914 | 0.13 |
ENSMUST00000060442.14
|
Gpr85
|
G protein-coupled receptor 85 |
| chr11_+_115656246 | 0.13 |
ENSMUST00000093912.11
ENSMUST00000136720.8 ENSMUST00000103034.10 ENSMUST00000141871.8 |
Tmem94
|
transmembrane protein 94 |
| chr10_-_128204545 | 0.12 |
ENSMUST00000220027.2
|
Coq10a
|
coenzyme Q10A |
| chr3_+_68375495 | 0.12 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr6_+_34840057 | 0.12 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr5_+_35156454 | 0.12 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr7_+_27430823 | 0.12 |
ENSMUST00000130997.8
ENSMUST00000042641.14 |
Zfp60
|
zinc finger protein 60 |
| chrX_+_7656225 | 0.11 |
ENSMUST00000136930.8
ENSMUST00000115675.9 ENSMUST00000101694.10 |
Gripap1
|
GRIP1 associated protein 1 |
| chr2_-_126517143 | 0.11 |
ENSMUST00000124972.8
|
Gabpb1
|
GA repeat binding protein, beta 1 |
| chr1_+_60948149 | 0.10 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chrX_+_106299484 | 0.10 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chrX_-_7054952 | 0.10 |
ENSMUST00000004428.14
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr1_-_160040286 | 0.10 |
ENSMUST00000195654.2
ENSMUST00000014370.11 |
Cacybp
|
calcyclin binding protein |
| chr16_-_22258320 | 0.10 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr2_-_126517383 | 0.10 |
ENSMUST00000103226.10
ENSMUST00000110424.9 |
Gabpb1
|
GA repeat binding protein, beta 1 |
| chr5_-_34345014 | 0.09 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr11_+_69909659 | 0.08 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr10_-_120815232 | 0.08 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr7_-_126522014 | 0.08 |
ENSMUST00000134134.3
ENSMUST00000119781.8 ENSMUST00000121612.4 |
Tmem219
|
transmembrane protein 219 |
| chr8_+_22717338 | 0.08 |
ENSMUST00000160585.2
ENSMUST00000162447.2 |
Thsd1
|
thrombospondin, type I, domain 1 |
| chr4_-_120808820 | 0.08 |
ENSMUST00000106280.8
|
Zfp69
|
zinc finger protein 69 |
| chr16_+_17051423 | 0.07 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chrX_-_93585668 | 0.07 |
ENSMUST00000026142.8
|
Maged1
|
MAGE family member D1 |
| chr7_+_27430796 | 0.07 |
ENSMUST00000108336.8
|
Zfp60
|
zinc finger protein 60 |
| chr10_+_115854118 | 0.07 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr16_-_43836681 | 0.07 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr2_-_17465410 | 0.06 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr5_+_20112500 | 0.06 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr18_+_61688329 | 0.06 |
ENSMUST00000165123.8
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr4_-_140780972 | 0.06 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr14_-_62998561 | 0.06 |
ENSMUST00000053959.7
ENSMUST00000223585.2 |
Ints6
|
integrator complex subunit 6 |
| chr8_+_79755194 | 0.06 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr3_+_103739877 | 0.06 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr1_+_178015287 | 0.05 |
ENSMUST00000159284.2
|
Desi2
|
desumoylating isopeptidase 2 |
| chr2_+_90770742 | 0.05 |
ENSMUST00000005643.14
ENSMUST00000111451.10 ENSMUST00000177642.8 ENSMUST00000068726.13 ENSMUST00000068747.14 |
Celf1
|
CUGBP, Elav-like family member 1 |
| chr14_+_76741625 | 0.05 |
ENSMUST00000177207.2
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr13_-_35147771 | 0.05 |
ENSMUST00000164155.2
ENSMUST00000021853.12 |
Eci3
|
enoyl-Coenzyme A delta isomerase 3 |
| chr5_+_35074862 | 0.05 |
ENSMUST00000202205.2
|
Msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr2_+_83554868 | 0.04 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chr2_+_127750978 | 0.04 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr18_+_35695485 | 0.04 |
ENSMUST00000235199.2
ENSMUST00000237744.2 ENSMUST00000236276.2 |
Matr3
|
matrin 3 |
| chr7_-_29935150 | 0.04 |
ENSMUST00000189482.2
|
Ovol3
|
ovo like zinc finger 3 |
| chr5_+_88731386 | 0.04 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr8_+_79754980 | 0.04 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr18_+_33072194 | 0.04 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr2_-_25471703 | 0.03 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr7_+_36397426 | 0.03 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr9_-_39484610 | 0.03 |
ENSMUST00000079178.3
|
Olfr959
|
olfactory receptor 959 |
| chr3_-_110158280 | 0.03 |
ENSMUST00000190378.2
ENSMUST00000106567.2 |
Prmt6
|
protein arginine N-methyltransferase 6 |
| chr1_+_60948307 | 0.03 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr7_+_83234118 | 0.03 |
ENSMUST00000039317.14
ENSMUST00000164944.2 |
Tmc3
|
transmembrane channel-like gene family 3 |
| chr14_+_20724366 | 0.03 |
ENSMUST00000048657.10
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr8_-_24928953 | 0.03 |
ENSMUST00000052622.6
|
Tcim
|
transcriptional and immune response regulator |
| chr5_+_20112704 | 0.03 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_-_55171699 | 0.03 |
ENSMUST00000144711.9
|
Wdr17
|
WD repeat domain 17 |
| chrX_-_142716085 | 0.03 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr2_+_65451100 | 0.02 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr1_-_75293997 | 0.02 |
ENSMUST00000189282.3
ENSMUST00000191254.7 |
Dnpep
|
aspartyl aminopeptidase |
| chrX_-_133012457 | 0.02 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr6_+_142244145 | 0.02 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr11_+_67167950 | 0.02 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr14_+_67470884 | 0.02 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr19_+_39102342 | 0.02 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr9_-_16289527 | 0.02 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr18_+_37952556 | 0.02 |
ENSMUST00000055935.11
|
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr6_-_68887957 | 0.02 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr16_+_84312257 | 0.02 |
ENSMUST00000210306.2
|
A730009L09Rik
|
RIKEN cDNA A730009L09 gene |
| chr15_-_76906832 | 0.02 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr2_-_160950936 | 0.02 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr1_+_179938904 | 0.02 |
ENSMUST00000145181.2
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr10_+_119655294 | 0.02 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr19_-_45619559 | 0.02 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr13_-_92931317 | 0.02 |
ENSMUST00000022213.8
|
Thbs4
|
thrombospondin 4 |
| chr14_+_102078038 | 0.02 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chr3_-_30563831 | 0.02 |
ENSMUST00000173495.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr7_-_109330915 | 0.02 |
ENSMUST00000035372.3
|
Ascl3
|
achaete-scute family bHLH transcription factor 3 |
| chr4_+_98919183 | 0.01 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr10_+_90412432 | 0.01 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_+_37952596 | 0.01 |
ENSMUST00000193890.2
ENSMUST00000193941.2 |
Pcdhgc5
|
protocadherin gamma subfamily C, 5 |
| chr6_-_93890520 | 0.01 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_+_90412570 | 0.01 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_-_89237422 | 0.01 |
ENSMUST00000099781.5
|
Olfr1238
|
olfactory receptor 1238 |
| chr13_-_12479804 | 0.01 |
ENSMUST00000124888.8
|
Lgals8
|
lectin, galactose binding, soluble 8 |
| chrX_+_41238193 | 0.01 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr2_-_140513382 | 0.01 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr10_+_90412638 | 0.01 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_+_19173246 | 0.01 |
ENSMUST00000037294.8
|
Tfap2d
|
transcription factor AP-2, delta |
| chr6_+_15185399 | 0.01 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr2_-_140231618 | 0.01 |
ENSMUST00000122367.8
ENSMUST00000120133.2 |
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr10_+_90412539 | 0.01 |
ENSMUST00000182284.8
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr14_+_67470735 | 0.01 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr6_+_141470105 | 0.01 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr5_-_69699965 | 0.01 |
ENSMUST00000031045.10
|
Yipf7
|
Yip1 domain family, member 7 |
| chr16_+_84312009 | 0.01 |
ENSMUST00000209558.2
|
A730009L09Rik
|
RIKEN cDNA A730009L09 gene |
| chr2_-_140513320 | 0.01 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr9_+_7628201 | 0.01 |
ENSMUST00000034487.4
|
Mmp20
|
matrix metallopeptidase 20 (enamelysin) |
| chr10_+_90412114 | 0.01 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr19_+_22670134 | 0.01 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr14_+_123897383 | 0.01 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chr4_+_119052476 | 0.01 |
ENSMUST00000030395.9
|
Svbp
|
small vasohibin binding protein |
| chr18_+_61688378 | 0.00 |
ENSMUST00000165721.8
ENSMUST00000115246.9 ENSMUST00000166990.8 ENSMUST00000163205.8 ENSMUST00000170862.8 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr3_+_40594899 | 0.00 |
ENSMUST00000091186.7
|
Intu
|
inturned planar cell polarity protein |
| chr1_-_164988342 | 0.00 |
ENSMUST00000027859.12
|
Tbx19
|
T-box 19 |
| chr14_+_102077937 | 0.00 |
ENSMUST00000159026.8
|
Lmo7
|
LIM domain only 7 |
| chr3_+_75981577 | 0.00 |
ENSMUST00000038364.15
|
Fstl5
|
follistatin-like 5 |
| chr14_-_34310438 | 0.00 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr6_+_141470261 | 0.00 |
ENSMUST00000203140.2
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr6_+_104469751 | 0.00 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chr5_-_69699932 | 0.00 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 1.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.2 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 | 0.5 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 0.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.5 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.5 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.8 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 0.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.3 | GO:0036233 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 0.1 | 0.3 | GO:0006507 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.5 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.2 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 1.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.9 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 1.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) regulation of protein localization to cilium(GO:1903564) positive regulation of protein localization to cilium(GO:1903566) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 1.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.5 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.2 | 0.5 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.1 | 1.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.3 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 0.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.8 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0048273 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |