Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
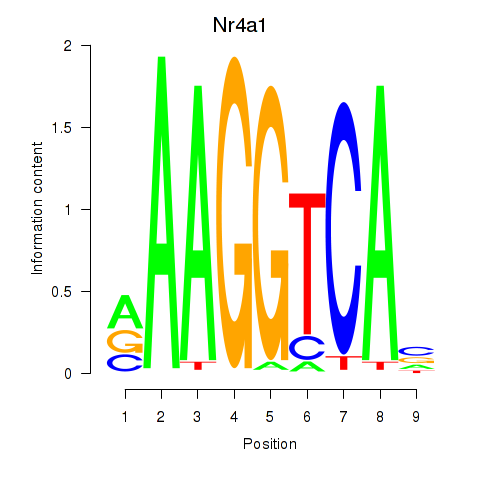
Results for Nr2f1_Nr4a1
Z-value: 0.69
Transcription factors associated with Nr2f1_Nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f1
|
ENSMUSG00000069171.15 | nuclear receptor subfamily 2, group F, member 1 |
|
Nr4a1
|
ENSMUSG00000023034.8 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f1 | mm39_v1_chr13_-_78347876_78347982 | -0.41 | 5.0e-01 | Click! |
| Nr4a1 | mm39_v1_chr15_+_101164719_101164740 | 0.22 | 7.2e-01 | Click! |
Activity profile of Nr2f1_Nr4a1 motif
Sorted Z-values of Nr2f1_Nr4a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_137548004 | 0.41 |
ENSMUST00000100841.9
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr10_-_78300802 | 0.37 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr7_+_125202653 | 0.33 |
ENSMUST00000206103.2
ENSMUST00000033000.8 |
Il21r
|
interleukin 21 receptor |
| chr7_-_100164007 | 0.29 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr7_-_79115915 | 0.25 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr16_-_20245138 | 0.24 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_+_14951478 | 0.23 |
ENSMUST00000029078.9
|
Car2
|
carbonic anhydrase 2 |
| chr5_+_135216090 | 0.22 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr11_+_23615612 | 0.22 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr17_-_56891576 | 0.22 |
ENSMUST00000075510.12
|
Safb2
|
scaffold attachment factor B2 |
| chr9_+_107957621 | 0.21 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_-_109363406 | 0.21 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr3_-_79535966 | 0.21 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr11_+_70548022 | 0.20 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr9_+_107957640 | 0.19 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr18_-_35760260 | 0.18 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr16_+_57173456 | 0.18 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_-_76474767 | 0.17 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr19_+_3372296 | 0.17 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr5_-_110596387 | 0.17 |
ENSMUST00000198768.3
|
Fbrsl1
|
fibrosin-like 1 |
| chr7_+_83281167 | 0.16 |
ENSMUST00000075418.15
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr9_+_108368032 | 0.16 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr18_+_60880149 | 0.16 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr7_-_4448631 | 0.16 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr11_+_68979332 | 0.16 |
ENSMUST00000117780.2
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr7_-_126074256 | 0.16 |
ENSMUST00000205440.2
ENSMUST00000032978.8 ENSMUST00000205340.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr16_+_3854806 | 0.16 |
ENSMUST00000137748.8
ENSMUST00000006136.11 ENSMUST00000157044.8 ENSMUST00000120009.8 |
Dnase1
|
deoxyribonuclease I |
| chr3_+_106393348 | 0.16 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr3_+_14951264 | 0.16 |
ENSMUST00000192609.6
|
Car2
|
carbonic anhydrase 2 |
| chr12_+_4132567 | 0.15 |
ENSMUST00000020986.15
ENSMUST00000049584.6 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr8_-_85663976 | 0.15 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr15_+_100659622 | 0.15 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr6_-_116693849 | 0.15 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr5_+_137517140 | 0.15 |
ENSMUST00000031727.10
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr8_+_85696396 | 0.15 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr1_+_173093568 | 0.14 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr7_+_130467564 | 0.14 |
ENSMUST00000075181.11
ENSMUST00000151119.9 ENSMUST00000048180.12 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr6_+_113259262 | 0.14 |
ENSMUST00000041203.6
|
Cpne9
|
copine family member IX |
| chr10_-_42354482 | 0.14 |
ENSMUST00000041024.15
|
Afg1l
|
AFG1 like ATPase |
| chr4_-_141450710 | 0.13 |
ENSMUST00000102484.5
ENSMUST00000177592.2 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr5_+_136112817 | 0.13 |
ENSMUST00000100570.10
|
Rasa4
|
RAS p21 protein activator 4 |
| chr16_+_32427738 | 0.13 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr4_-_119047202 | 0.13 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr2_-_25911691 | 0.13 |
ENSMUST00000036509.14
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr19_-_43512929 | 0.13 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr1_-_160079007 | 0.13 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr17_-_46343291 | 0.13 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr17_+_31120785 | 0.13 |
ENSMUST00000114574.3
|
Glp1r
|
glucagon-like peptide 1 receptor |
| chr13_+_54346116 | 0.13 |
ENSMUST00000038101.4
|
Hrh2
|
histamine receptor H2 |
| chr1_-_36408254 | 0.13 |
ENSMUST00000010597.10
|
Kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr17_+_29579882 | 0.12 |
ENSMUST00000024810.8
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr8_+_85696453 | 0.12 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr5_-_31359559 | 0.12 |
ENSMUST00000202929.2
ENSMUST00000201231.2 ENSMUST00000114590.8 |
Zfp513
|
zinc finger protein 513 |
| chr1_-_36408202 | 0.12 |
ENSMUST00000185912.7
|
Kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr5_+_136112765 | 0.12 |
ENSMUST00000042135.14
|
Rasa4
|
RAS p21 protein activator 4 |
| chr16_-_43800109 | 0.12 |
ENSMUST00000231700.2
|
Zdhhc23
|
zinc finger, DHHC domain containing 23 |
| chr11_+_97697328 | 0.12 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr2_-_167334746 | 0.12 |
ENSMUST00000109211.9
ENSMUST00000057627.16 |
Spata2
|
spermatogenesis associated 2 |
| chr5_+_137516810 | 0.12 |
ENSMUST00000197624.5
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr4_-_130068484 | 0.12 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chrX_-_106859842 | 0.12 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr7_-_143102524 | 0.12 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr17_-_36220924 | 0.12 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr19_+_5742880 | 0.12 |
ENSMUST00000235661.2
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr13_-_64396422 | 0.12 |
ENSMUST00000109769.10
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr14_+_54862762 | 0.12 |
ENSMUST00000097177.5
|
Psmb11
|
proteasome (prosome, macropain) subunit, beta type, 11 |
| chr16_+_32427789 | 0.12 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr6_+_21985902 | 0.12 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr17_+_30845909 | 0.12 |
ENSMUST00000236140.2
ENSMUST00000236118.2 ENSMUST00000235390.2 |
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr5_-_108822619 | 0.12 |
ENSMUST00000119270.2
ENSMUST00000163328.8 ENSMUST00000136227.2 |
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr7_-_126074222 | 0.12 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr7_+_79674562 | 0.12 |
ENSMUST00000164056.9
ENSMUST00000166250.8 |
Zfp710
|
zinc finger protein 710 |
| chr17_+_5542832 | 0.11 |
ENSMUST00000089185.6
|
Zdhhc14
|
zinc finger, DHHC domain containing 14 |
| chr13_-_21586858 | 0.11 |
ENSMUST00000117721.8
ENSMUST00000070785.16 ENSMUST00000116433.2 ENSMUST00000223831.2 ENSMUST00000116434.11 ENSMUST00000224820.2 |
Zkscan3
|
zinc finger with KRAB and SCAN domains 3 |
| chr11_-_76468527 | 0.11 |
ENSMUST00000176179.8
|
Abr
|
active BCR-related gene |
| chr4_-_119047167 | 0.11 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr12_-_100865783 | 0.11 |
ENSMUST00000053668.10
|
Gpr68
|
G protein-coupled receptor 68 |
| chr19_-_7218363 | 0.11 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr1_-_175520185 | 0.11 |
ENSMUST00000104984.4
ENSMUST00000209720.2 ENSMUST00000211489.2 ENSMUST00000210367.2 ENSMUST00000027809.8 |
Chml
Opn3
|
choroideremia-like opsin 3 |
| chr19_-_6899173 | 0.11 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr7_-_79115760 | 0.11 |
ENSMUST00000125562.2
|
Polg
|
polymerase (DNA directed), gamma |
| chr8_+_85696695 | 0.11 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr19_+_4806544 | 0.11 |
ENSMUST00000182821.8
ENSMUST00000036744.8 |
Rbm4b
|
RNA binding motif protein 4B |
| chr9_-_43027809 | 0.11 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chrX_-_149372840 | 0.11 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr16_+_91526169 | 0.11 |
ENSMUST00000114001.8
ENSMUST00000113999.8 ENSMUST00000064797.12 ENSMUST00000114002.9 ENSMUST00000095909.10 ENSMUST00000056482.14 ENSMUST00000113996.8 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr19_+_43770619 | 0.11 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr7_-_46365108 | 0.11 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr2_-_156921135 | 0.10 |
ENSMUST00000069098.7
|
Soga1
|
suppressor of glucose, autophagy associated 1 |
| chr2_+_103900164 | 0.10 |
ENSMUST00000111131.9
ENSMUST00000111132.8 ENSMUST00000129749.8 |
Cd59b
|
CD59b antigen |
| chr19_+_47079187 | 0.10 |
ENSMUST00000072141.4
|
Pdcd11
|
programmed cell death 11 |
| chr1_-_45964730 | 0.10 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr5_-_90487583 | 0.10 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr7_-_4525426 | 0.10 |
ENSMUST00000209148.2
ENSMUST00000098859.10 |
Tnni3
|
troponin I, cardiac 3 |
| chr9_+_118335294 | 0.10 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chrX_+_10351360 | 0.10 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr11_+_90528953 | 0.10 |
ENSMUST00000020851.15
|
Cox11
|
cytochrome c oxidase assembly protein 11, copper chaperone |
| chr5_-_92475927 | 0.10 |
ENSMUST00000113093.5
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr2_+_79538124 | 0.10 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr15_-_102097387 | 0.10 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr4_-_118266416 | 0.10 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr7_-_16761732 | 0.10 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr1_-_134731516 | 0.10 |
ENSMUST00000238280.2
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_+_99812912 | 0.10 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr3_-_59118293 | 0.10 |
ENSMUST00000040622.3
|
P2ry13
|
purinergic receptor P2Y, G-protein coupled 13 |
| chr1_-_172125555 | 0.10 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr2_-_11782381 | 0.10 |
ENSMUST00000071564.14
|
Fbh1
|
F-box DNA helicase 1 |
| chr4_+_107659474 | 0.10 |
ENSMUST00000106733.10
ENSMUST00000238651.2 ENSMUST00000030356.10 ENSMUST00000238421.2 ENSMUST00000126573.8 ENSMUST00000238569.2 ENSMUST00000106732.10 |
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr1_-_190950129 | 0.10 |
ENSMUST00000195117.2
|
Atf3
|
activating transcription factor 3 |
| chr4_-_149822480 | 0.10 |
ENSMUST00000105687.9
ENSMUST00000054459.11 ENSMUST00000103208.2 |
Tmem201
|
transmembrane protein 201 |
| chr1_-_63215952 | 0.10 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr2_-_136733282 | 0.10 |
ENSMUST00000028730.13
ENSMUST00000110089.9 ENSMUST00000227806.2 |
Mkks
ENSMUSG00000115423.3
|
McKusick-Kaufman syndrome novel protein |
| chr13_+_84370405 | 0.10 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr9_+_108367801 | 0.09 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr7_-_4448180 | 0.09 |
ENSMUST00000138798.2
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr15_-_89310060 | 0.09 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr8_-_121394860 | 0.09 |
ENSMUST00000034277.14
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr16_+_17149235 | 0.09 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr11_+_70548622 | 0.09 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr9_-_42383494 | 0.09 |
ENSMUST00000128959.8
ENSMUST00000066148.12 ENSMUST00000138506.8 |
Tbcel
|
tubulin folding cofactor E-like |
| chr7_-_143102792 | 0.09 |
ENSMUST00000072727.7
ENSMUST00000207948.2 ENSMUST00000208190.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr10_-_90959817 | 0.09 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr1_-_93682490 | 0.09 |
ENSMUST00000190116.7
|
Thap4
|
THAP domain containing 4 |
| chr10_+_4561974 | 0.09 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr2_-_127324419 | 0.09 |
ENSMUST00000088538.6
|
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr15_+_84553801 | 0.09 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr6_+_41515152 | 0.09 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chrX_+_159551009 | 0.09 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr15_+_76227695 | 0.09 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr16_-_8455525 | 0.09 |
ENSMUST00000052505.10
|
Tmem186
|
transmembrane protein 186 |
| chr11_+_6242119 | 0.09 |
ENSMUST00000135124.8
|
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr14_+_79086665 | 0.09 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr6_+_113508636 | 0.09 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr12_-_84240781 | 0.09 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr4_+_97665992 | 0.09 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr6_-_142910094 | 0.09 |
ENSMUST00000032421.4
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr13_-_74498320 | 0.08 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr8_-_13544478 | 0.08 |
ENSMUST00000033828.7
|
Gas6
|
growth arrest specific 6 |
| chr8_-_106198112 | 0.08 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr7_+_51537645 | 0.08 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr2_-_131016573 | 0.08 |
ENSMUST00000127987.2
|
Spef1
|
sperm flagellar 1 |
| chr13_+_92981257 | 0.08 |
ENSMUST00000076169.4
|
Mtx3
|
metaxin 3 |
| chr15_+_86098660 | 0.08 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr5_+_107645626 | 0.08 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr15_-_102097466 | 0.08 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr6_-_91450486 | 0.08 |
ENSMUST00000040835.9
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr2_-_25911544 | 0.08 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chrX_+_55493325 | 0.08 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr14_-_21898992 | 0.08 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr15_-_79976016 | 0.08 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr4_+_131600918 | 0.08 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr5_-_25200745 | 0.08 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr16_+_10363254 | 0.08 |
ENSMUST00000115827.8
ENSMUST00000150894.2 ENSMUST00000038145.13 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr8_+_121395047 | 0.08 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr5_+_137628377 | 0.08 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr4_+_107659361 | 0.08 |
ENSMUST00000106731.4
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr10_-_42354276 | 0.08 |
ENSMUST00000151747.8
|
Afg1l
|
AFG1 like ATPase |
| chr1_-_74544946 | 0.08 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr4_+_97665843 | 0.08 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr17_+_29251602 | 0.08 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr9_+_64142483 | 0.08 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr1_-_172034251 | 0.07 |
ENSMUST00000155109.2
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr9_+_39932760 | 0.07 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr5_-_31359276 | 0.07 |
ENSMUST00000031562.11
|
Zfp513
|
zinc finger protein 513 |
| chr7_+_28533279 | 0.07 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr6_-_125213911 | 0.07 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr16_-_23339548 | 0.07 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr14_+_19801333 | 0.07 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr14_+_79086492 | 0.07 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr11_-_99134885 | 0.07 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr2_+_58991182 | 0.07 |
ENSMUST00000168631.8
ENSMUST00000102754.11 ENSMUST00000123908.8 |
Pkp4
|
plakophilin 4 |
| chr8_+_27575611 | 0.07 |
ENSMUST00000178514.8
ENSMUST00000033876.14 |
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr11_-_94187893 | 0.07 |
ENSMUST00000132623.9
|
Luc7l3
|
LUC7-like 3 (S. cerevisiae) |
| chr7_-_126944754 | 0.07 |
ENSMUST00000205266.2
|
Zfp768
|
zinc finger protein 768 |
| chr4_+_118266582 | 0.07 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr7_-_30755007 | 0.07 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr16_-_20245071 | 0.07 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_+_87754310 | 0.07 |
ENSMUST00000029709.7
|
Sh2d2a
|
SH2 domain containing 2A |
| chr14_-_50390356 | 0.07 |
ENSMUST00000215451.2
ENSMUST00000213163.2 ENSMUST00000215327.2 |
Olfr729
|
olfactory receptor 729 |
| chr2_+_153133326 | 0.07 |
ENSMUST00000028977.7
|
Kif3b
|
kinesin family member 3B |
| chr9_-_44352312 | 0.07 |
ENSMUST00000215980.2
ENSMUST00000098837.3 |
Foxr1
|
forkhead box R1 |
| chr10_-_119075910 | 0.07 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr14_+_79753055 | 0.07 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr5_-_110596352 | 0.07 |
ENSMUST00000069483.12
ENSMUST00000200293.2 |
Fbrsl1
|
fibrosin-like 1 |
| chr15_-_102425241 | 0.07 |
ENSMUST00000169162.8
ENSMUST00000023812.10 ENSMUST00000165174.8 ENSMUST00000169367.8 ENSMUST00000169377.8 |
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr5_-_35897331 | 0.07 |
ENSMUST00000201511.2
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr1_+_88154727 | 0.07 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr13_+_74498418 | 0.07 |
ENSMUST00000022063.14
|
Ccdc127
|
coiled-coil domain containing 127 |
| chr6_-_69584812 | 0.07 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr14_-_20844074 | 0.06 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_-_63215812 | 0.06 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr4_-_45408646 | 0.06 |
ENSMUST00000153904.2
ENSMUST00000132815.3 ENSMUST00000107796.8 ENSMUST00000116341.4 |
Slc25a51
|
solute carrier family 25, member 51 |
| chr11_-_61384998 | 0.06 |
ENSMUST00000101085.9
ENSMUST00000079080.13 ENSMUST00000108714.2 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr12_-_100874440 | 0.06 |
ENSMUST00000110065.8
ENSMUST00000110066.8 |
Gpr68
|
G protein-coupled receptor 68 |
| chr12_-_110945415 | 0.06 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr17_-_25492129 | 0.06 |
ENSMUST00000173231.2
|
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr10_+_67021509 | 0.06 |
ENSMUST00000173689.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr7_-_28297565 | 0.06 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr8_+_48563038 | 0.06 |
ENSMUST00000033966.13
|
Dctd
|
dCMP deaminase |
| chr1_-_169796709 | 0.06 |
ENSMUST00000027989.13
ENSMUST00000111353.4 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f1_Nr4a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0090088 | positive regulation of cellular pH reduction(GO:0032849) dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.4 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.1 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.0 | 0.1 | GO:0039663 | positive regulation of glomerular filtration(GO:0003104) fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.1 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.4 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.0 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.1 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.0 | GO:1904346 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:1903273 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.0 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 0.3 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.1 | 0.2 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.1 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0050354 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.0 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.0 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |