Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
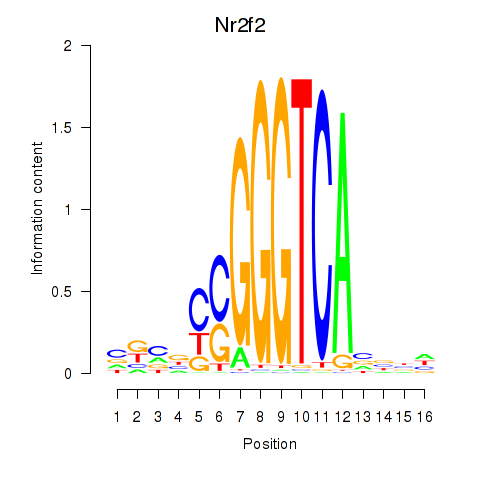
Results for Nr2f2
Z-value: 0.64
Transcription factors associated with Nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f2
|
ENSMUSG00000030551.15 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f2 | mm39_v1_chr7_-_70009669_70009697 | -0.87 | 5.3e-02 | Click! |
Activity profile of Nr2f2 motif
Sorted Z-values of Nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_11747721 | 0.39 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr18_+_60907698 | 0.39 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chrX_+_93679671 | 0.36 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr2_-_77110933 | 0.32 |
ENSMUST00000102659.2
|
Sestd1
|
SEC14 and spectrin domains 1 |
| chr18_+_60907668 | 0.31 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr11_-_120687195 | 0.26 |
ENSMUST00000143139.8
ENSMUST00000129955.2 ENSMUST00000026151.11 ENSMUST00000167023.8 ENSMUST00000106133.8 ENSMUST00000106135.8 |
Dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr11_-_101357046 | 0.26 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chr6_-_24527545 | 0.23 |
ENSMUST00000118558.5
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr4_+_127137577 | 0.22 |
ENSMUST00000142029.2
|
Smim12
|
small integral membrane protein 12 |
| chr1_+_181952302 | 0.22 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr19_-_6886898 | 0.21 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr13_+_73476629 | 0.21 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr19_+_21630887 | 0.21 |
ENSMUST00000052556.5
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr2_+_18681812 | 0.20 |
ENSMUST00000028071.13
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr7_+_18883647 | 0.20 |
ENSMUST00000049294.4
|
Snrpd2
|
small nuclear ribonucleoprotein D2 |
| chr15_+_84076423 | 0.20 |
ENSMUST00000023071.8
|
Samm50
|
SAMM50 sorting and assembly machinery component |
| chr8_+_53964721 | 0.20 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr2_+_127112127 | 0.20 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr3_+_100829798 | 0.19 |
ENSMUST00000106980.9
|
Trim45
|
tripartite motif-containing 45 |
| chr2_-_151586063 | 0.19 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr1_+_160022785 | 0.19 |
ENSMUST00000135680.8
ENSMUST00000097193.3 |
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr17_-_26014613 | 0.19 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chr5_+_35106778 | 0.19 |
ENSMUST00000030984.14
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr11_-_106890307 | 0.18 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr10_+_79889863 | 0.18 |
ENSMUST00000097227.11
|
Gpx4
|
glutathione peroxidase 4 |
| chr1_+_91468266 | 0.18 |
ENSMUST00000086843.11
|
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chrX_+_10581248 | 0.18 |
ENSMUST00000144356.8
|
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr9_-_44318597 | 0.18 |
ENSMUST00000217163.2
|
Trappc4
|
trafficking protein particle complex 4 |
| chr1_+_127132712 | 0.17 |
ENSMUST00000038361.11
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr17_+_8384333 | 0.17 |
ENSMUST00000097419.10
ENSMUST00000024636.15 |
Cep43
|
centrosomal protein 43 |
| chr3_+_100829750 | 0.17 |
ENSMUST00000037409.13
|
Trim45
|
tripartite motif-containing 45 |
| chr13_-_96807346 | 0.16 |
ENSMUST00000022176.15
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr9_-_44318823 | 0.16 |
ENSMUST00000034623.8
|
Trappc4
|
trafficking protein particle complex 4 |
| chr5_-_117425648 | 0.16 |
ENSMUST00000111973.8
ENSMUST00000036951.13 |
Pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr14_+_62529924 | 0.16 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr11_-_40583493 | 0.16 |
ENSMUST00000040167.11
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr5_+_65264863 | 0.15 |
ENSMUST00000204097.3
|
Klhl5
|
kelch-like 5 |
| chr17_+_6130205 | 0.15 |
ENSMUST00000100955.3
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr17_+_28075415 | 0.15 |
ENSMUST00000114849.3
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr6_-_128414616 | 0.15 |
ENSMUST00000151796.3
|
Fkbp4
|
FK506 binding protein 4 |
| chr6_-_126826077 | 0.15 |
ENSMUST00000205002.3
ENSMUST00000088194.7 |
Ndufa9
|
NADH:ubiquinone oxidoreductase subunit A9 |
| chr2_+_150590956 | 0.15 |
ENSMUST00000094467.6
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr5_-_110987604 | 0.15 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr19_+_32463151 | 0.14 |
ENSMUST00000025827.10
|
Minpp1
|
multiple inositol polyphosphate histidine phosphatase 1 |
| chr10_-_80742244 | 0.14 |
ENSMUST00000105332.3
|
Lmnb2
|
lamin B2 |
| chr8_-_96615138 | 0.14 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr7_-_26866157 | 0.14 |
ENSMUST00000080058.11
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr8_+_71047110 | 0.14 |
ENSMUST00000019283.10
ENSMUST00000210005.2 |
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr2_-_155668567 | 0.14 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr9_+_107765320 | 0.14 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr19_-_6887361 | 0.14 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr4_+_135691030 | 0.14 |
ENSMUST00000102541.10
ENSMUST00000149636.2 ENSMUST00000143304.2 |
Gale
|
galactose-4-epimerase, UDP |
| chr1_+_91468796 | 0.14 |
ENSMUST00000188081.7
ENSMUST00000188879.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr11_+_78192355 | 0.14 |
ENSMUST00000045026.4
|
Spag5
|
sperm associated antigen 5 |
| chr4_+_152123772 | 0.14 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr2_+_163500290 | 0.14 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr11_+_97576724 | 0.13 |
ENSMUST00000107583.3
|
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr4_+_139350152 | 0.13 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr14_-_31552335 | 0.13 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr19_+_56385531 | 0.13 |
ENSMUST00000026062.10
|
Casp7
|
caspase 7 |
| chr4_-_132056725 | 0.13 |
ENSMUST00000127402.8
ENSMUST00000105962.10 ENSMUST00000030730.14 ENSMUST00000105960.3 |
Trnau1ap
|
tRNA selenocysteine 1 associated protein 1 |
| chr12_-_101942172 | 0.13 |
ENSMUST00000221191.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr2_+_36026395 | 0.13 |
ENSMUST00000028250.9
|
Mrrf
|
mitochondrial ribosome recycling factor |
| chr4_+_24898074 | 0.12 |
ENSMUST00000029925.10
ENSMUST00000151249.2 |
Ndufaf4
|
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
| chr11_+_115705550 | 0.12 |
ENSMUST00000021134.10
ENSMUST00000106481.9 |
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chr15_-_4008913 | 0.12 |
ENSMUST00000022791.9
|
Fbxo4
|
F-box protein 4 |
| chr12_-_69243871 | 0.12 |
ENSMUST00000223192.2
|
Gm49383
|
predicted gene, 49383 |
| chr5_+_114582327 | 0.12 |
ENSMUST00000137167.8
ENSMUST00000112239.9 ENSMUST00000124260.8 ENSMUST00000125650.6 ENSMUST00000043760.15 |
Mvk
|
mevalonate kinase |
| chr5_+_143534455 | 0.12 |
ENSMUST00000169329.8
ENSMUST00000067145.12 ENSMUST00000119488.2 ENSMUST00000118121.2 ENSMUST00000200267.2 ENSMUST00000196487.2 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr1_-_91340884 | 0.12 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr19_-_6886965 | 0.12 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr6_-_119365632 | 0.12 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr12_+_72807985 | 0.12 |
ENSMUST00000021514.10
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform |
| chr1_+_180731843 | 0.12 |
ENSMUST00000027802.9
|
Pycr2
|
pyrroline-5-carboxylate reductase family, member 2 |
| chr8_-_85414220 | 0.12 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr6_-_120508250 | 0.12 |
ENSMUST00000075303.7
|
Hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr13_-_38842967 | 0.12 |
ENSMUST00000001757.9
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr6_+_122285615 | 0.12 |
ENSMUST00000007602.15
ENSMUST00000112610.2 |
M6pr
|
mannose-6-phosphate receptor, cation dependent |
| chr7_-_141614758 | 0.11 |
ENSMUST00000211000.2
ENSMUST00000209725.2 ENSMUST00000084418.4 |
Mob2
|
MOB kinase activator 2 |
| chr3_+_94600863 | 0.11 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr19_-_44543838 | 0.11 |
ENSMUST00000167027.2
ENSMUST00000171415.8 ENSMUST00000026222.11 |
Ndufb8
|
NADH:ubiquinone oxidoreductase subunit B8 |
| chr9_+_119168714 | 0.11 |
ENSMUST00000176351.8
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr2_+_144398149 | 0.11 |
ENSMUST00000143573.8
ENSMUST00000028916.15 ENSMUST00000155258.2 |
Sec23b
|
SEC23 homolog B, COPII coat complex component |
| chr13_+_33187205 | 0.11 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr8_+_85786684 | 0.11 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr4_+_107736942 | 0.10 |
ENSMUST00000030348.6
|
Magoh
|
mago homolog, exon junction complex core component |
| chr8_-_47986473 | 0.10 |
ENSMUST00000039061.15
|
Trappc11
|
trafficking protein particle complex 11 |
| chr12_-_72711533 | 0.10 |
ENSMUST00000021512.11
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr4_-_86775602 | 0.10 |
ENSMUST00000102814.5
|
Rps6
|
ribosomal protein S6 |
| chr5_-_65694483 | 0.10 |
ENSMUST00000149167.2
|
Smim14
|
small integral membrane protein 14 |
| chr11_-_117764258 | 0.10 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr3_+_157239988 | 0.10 |
ENSMUST00000029831.16
ENSMUST00000106057.8 |
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr9_+_106158549 | 0.10 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr5_+_45650716 | 0.10 |
ENSMUST00000046122.11
|
Lap3
|
leucine aminopeptidase 3 |
| chr5_+_100946493 | 0.10 |
ENSMUST00000016977.15
ENSMUST00000112898.8 ENSMUST00000112901.2 |
Mrps18c
|
mitochondrial ribosomal protein S18C |
| chr19_+_21630540 | 0.10 |
ENSMUST00000235332.2
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr16_+_55786638 | 0.10 |
ENSMUST00000023269.5
|
Rpl24
|
ribosomal protein L24 |
| chr19_-_4978238 | 0.10 |
ENSMUST00000237394.2
ENSMUST00000025851.4 |
Dpp3
|
dipeptidylpeptidase 3 |
| chr10_+_127871444 | 0.10 |
ENSMUST00000073868.9
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr17_-_81372483 | 0.10 |
ENSMUST00000025093.6
|
Thumpd2
|
THUMP domain containing 2 |
| chr7_+_19144950 | 0.10 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr5_+_21629932 | 0.10 |
ENSMUST00000056045.5
|
Fam185a
|
family with sequence similarity 185, member A |
| chr7_+_138792890 | 0.10 |
ENSMUST00000016124.15
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr15_-_74671382 | 0.10 |
ENSMUST00000168815.8
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr1_+_91468409 | 0.10 |
ENSMUST00000027538.9
ENSMUST00000190484.7 ENSMUST00000186068.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr7_-_113853894 | 0.10 |
ENSMUST00000033012.9
|
Copb1
|
coatomer protein complex, subunit beta 1 |
| chr3_+_89979948 | 0.09 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr16_-_10360893 | 0.09 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr7_+_28533279 | 0.09 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr2_-_179915276 | 0.09 |
ENSMUST00000108891.2
|
Cables2
|
CDK5 and Abl enzyme substrate 2 |
| chr16_+_35892437 | 0.09 |
ENSMUST00000163352.9
ENSMUST00000231468.2 |
Ccdc58
|
coiled-coil domain containing 58 |
| chr3_-_68952030 | 0.09 |
ENSMUST00000136512.3
|
Trim59
|
tripartite motif-containing 59 |
| chr3_+_51324022 | 0.09 |
ENSMUST00000192419.6
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr1_+_172328768 | 0.09 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr5_+_45650821 | 0.09 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr11_-_100986192 | 0.09 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr7_+_4880111 | 0.09 |
ENSMUST00000125249.3
|
Isoc2a
|
isochorismatase domain containing 2a |
| chr19_-_42117420 | 0.09 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chr2_+_112092271 | 0.09 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr2_+_30306116 | 0.09 |
ENSMUST00000113601.10
ENSMUST00000113603.10 |
Ptpa
|
protein phosphatase 2 protein activator |
| chr5_-_114266228 | 0.09 |
ENSMUST00000053657.13
ENSMUST00000149418.2 ENSMUST00000112279.2 |
Alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr2_-_36026785 | 0.09 |
ENSMUST00000028251.10
|
Rbm18
|
RNA binding motif protein 18 |
| chr5_+_129097133 | 0.09 |
ENSMUST00000031383.14
ENSMUST00000111343.2 |
Ran
|
RAN, member RAS oncogene family |
| chr2_+_130266253 | 0.09 |
ENSMUST00000128994.8
ENSMUST00000028900.11 |
Vps16
|
VSP16 CORVET/HOPS core subunit |
| chr6_+_115398996 | 0.09 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr2_+_68966125 | 0.09 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr11_-_21521934 | 0.09 |
ENSMUST00000239073.2
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr17_-_71833752 | 0.09 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chr4_-_16013867 | 0.09 |
ENSMUST00000037198.10
|
Osgin2
|
oxidative stress induced growth inhibitor family member 2 |
| chr2_-_36026614 | 0.08 |
ENSMUST00000122456.8
|
Rbm18
|
RNA binding motif protein 18 |
| chr5_+_76677150 | 0.08 |
ENSMUST00000087133.11
ENSMUST00000113493.8 ENSMUST00000049469.13 |
Exoc1
|
exocyst complex component 1 |
| chr6_-_136804728 | 0.08 |
ENSMUST00000146348.4
|
Wbp11
|
WW domain binding protein 11 |
| chr13_-_73476561 | 0.08 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr2_+_144398226 | 0.08 |
ENSMUST00000155876.8
ENSMUST00000149697.3 |
Sec23b
|
SEC23 homolog B, COPII coat complex component |
| chr3_+_157239740 | 0.08 |
ENSMUST00000106058.8
|
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr11_+_83189831 | 0.08 |
ENSMUST00000176944.8
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr2_-_104847360 | 0.08 |
ENSMUST00000111110.3
ENSMUST00000028592.12 |
Eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr17_-_12726591 | 0.08 |
ENSMUST00000024595.4
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr4_+_53440389 | 0.08 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr11_-_117671436 | 0.08 |
ENSMUST00000026659.10
ENSMUST00000127227.2 |
Tmc6
|
transmembrane channel-like gene family 6 |
| chr17_+_6130061 | 0.08 |
ENSMUST00000039487.10
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr8_-_69636825 | 0.08 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr10_-_43416941 | 0.08 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr4_+_118266582 | 0.08 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr17_-_26004298 | 0.08 |
ENSMUST00000150324.8
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr3_-_88317601 | 0.08 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr12_+_101942222 | 0.08 |
ENSMUST00000047357.10
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr10_-_43777712 | 0.08 |
ENSMUST00000020012.7
|
Qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr8_-_124709859 | 0.08 |
ENSMUST00000075578.7
|
Abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr4_-_62443273 | 0.08 |
ENSMUST00000030091.10
|
Pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr12_+_111505253 | 0.08 |
ENSMUST00000220803.2
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr7_+_140972070 | 0.08 |
ENSMUST00000211654.2
ENSMUST00000026576.5 |
Taldo1
|
transaldolase 1 |
| chr12_+_111504450 | 0.07 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr10_-_30494333 | 0.07 |
ENSMUST00000019925.7
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr11_-_106890195 | 0.07 |
ENSMUST00000106768.2
ENSMUST00000144834.8 |
Kpna2
|
karyopherin (importin) alpha 2 |
| chr10_-_30476658 | 0.07 |
ENSMUST00000019927.7
|
Trmt11
|
tRNA methyltransferase 11 |
| chr13_-_96807326 | 0.07 |
ENSMUST00000169196.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr7_+_130247912 | 0.07 |
ENSMUST00000207549.2
ENSMUST00000209108.2 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chrX_-_133501874 | 0.07 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr8_+_84293300 | 0.07 |
ENSMUST00000036996.6
|
Ndufb7
|
NADH:ubiquinone oxidoreductase subunit B7 |
| chr11_-_21522193 | 0.07 |
ENSMUST00000102874.11
ENSMUST00000238916.2 |
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr6_+_59185846 | 0.07 |
ENSMUST00000062626.4
|
Tigd2
|
tigger transposable element derived 2 |
| chr11_-_97944239 | 0.07 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr9_+_122180673 | 0.07 |
ENSMUST00000156520.8
ENSMUST00000111497.5 |
Abhd5
|
abhydrolase domain containing 5 |
| chr8_-_72178340 | 0.07 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr6_-_134609350 | 0.07 |
ENSMUST00000047443.5
|
Mansc1
|
MANSC domain containing 1 |
| chr11_-_57409423 | 0.07 |
ENSMUST00000108850.2
ENSMUST00000020831.13 |
Fam114a2
|
family with sequence similarity 114, member A2 |
| chr4_+_129181407 | 0.07 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr7_-_45109075 | 0.07 |
ENSMUST00000210864.2
|
Ftl1
|
ferritin light polypeptide 1 |
| chr17_-_67661382 | 0.07 |
ENSMUST00000223982.2
ENSMUST00000224091.2 |
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr7_-_141614590 | 0.07 |
ENSMUST00000211206.2
|
Mob2
|
MOB kinase activator 2 |
| chr9_+_107464841 | 0.07 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr9_-_22043083 | 0.07 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr12_+_17594795 | 0.07 |
ENSMUST00000171737.3
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr2_+_179899166 | 0.07 |
ENSMUST00000059080.7
|
Rps21
|
ribosomal protein S21 |
| chr2_+_18677195 | 0.07 |
ENSMUST00000171845.8
ENSMUST00000061158.5 |
Commd3
|
COMM domain containing 3 |
| chr4_+_137989526 | 0.06 |
ENSMUST00000030539.10
|
Kif17
|
kinesin family member 17 |
| chr2_+_105734975 | 0.06 |
ENSMUST00000037499.6
|
Immp1l
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr10_-_84963841 | 0.06 |
ENSMUST00000214193.2
ENSMUST00000050813.4 ENSMUST00000217027.2 |
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr7_+_138793150 | 0.06 |
ENSMUST00000106104.8
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr9_+_75348800 | 0.06 |
ENSMUST00000048937.6
|
Leo1
|
Leo1, Paf1/RNA polymerase II complex component |
| chr2_+_118943274 | 0.06 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr13_+_21364330 | 0.06 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr19_+_37184927 | 0.06 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr18_+_76374959 | 0.06 |
ENSMUST00000171256.8
|
Smad2
|
SMAD family member 2 |
| chr4_+_152381662 | 0.06 |
ENSMUST00000048892.14
|
Icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr1_+_87332638 | 0.06 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr11_+_118367651 | 0.06 |
ENSMUST00000135383.9
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr17_+_29898221 | 0.06 |
ENSMUST00000129864.2
|
Cmtr1
|
cap methyltransferase 1 |
| chr2_-_60213639 | 0.06 |
ENSMUST00000112533.8
|
Ly75
|
lymphocyte antigen 75 |
| chr17_+_34823790 | 0.06 |
ENSMUST00000173242.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chrX_-_133501677 | 0.06 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr4_+_118266526 | 0.06 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr15_-_79326001 | 0.06 |
ENSMUST00000122044.8
ENSMUST00000135519.2 |
Csnk1e
|
casein kinase 1, epsilon |
| chr17_+_29312737 | 0.06 |
ENSMUST00000023829.8
ENSMUST00000233296.2 |
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr6_+_115339253 | 0.06 |
ENSMUST00000203732.3
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr11_-_70910058 | 0.06 |
ENSMUST00000108523.10
ENSMUST00000143850.8 |
Derl2
|
Der1-like domain family, member 2 |
| chr2_-_34716083 | 0.06 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr17_-_26463092 | 0.06 |
ENSMUST00000118487.8
ENSMUST00000234262.2 |
Fam234a
|
family with sequence similarity 234, member A |
| chr11_+_3939924 | 0.06 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.5 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.3 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.2 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.1 | 0.2 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.3 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.4 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.0 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.1 | 0.2 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.6 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |