Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nr3c2
Z-value: 1.10
Transcription factors associated with Nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr3c2
|
ENSMUSG00000031618.14 | nuclear receptor subfamily 3, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c2 | mm39_v1_chr8_+_77626400_77626456 | 0.90 | 3.8e-02 | Click! |
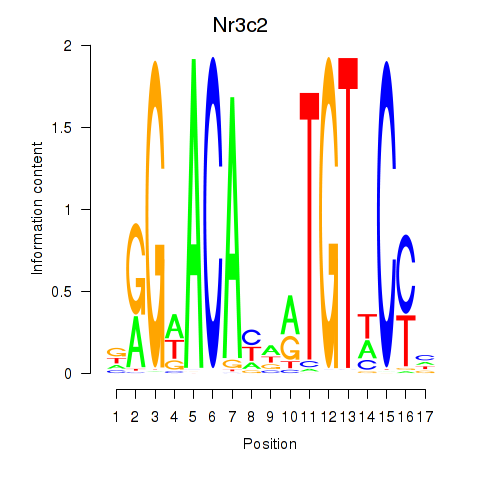
Activity profile of Nr3c2 motif
Sorted Z-values of Nr3c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_123195986 | 3.19 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr17_-_23531402 | 1.20 |
ENSMUST00000168033.3
|
Vmn2r114
|
vomeronasal 2, receptor 114 |
| chr11_+_68989763 | 1.08 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr5_+_123280250 | 0.82 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr14_+_14159978 | 0.82 |
ENSMUST00000137133.2
ENSMUST00000036070.15 ENSMUST00000121887.8 |
Fam107a
|
family with sequence similarity 107, member A |
| chr17_+_22819932 | 0.77 |
ENSMUST00000097381.5
ENSMUST00000234882.2 |
Vmn2r112
|
vomeronasal 2, receptor 112 |
| chr6_-_146403410 | 0.59 |
ENSMUST00000053273.15
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr14_+_35816874 | 0.59 |
ENSMUST00000226305.2
|
4930474N05Rik
|
RIKEN cDNA 4930474N05 gene |
| chr10_+_128061699 | 0.57 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr11_+_50905063 | 0.55 |
ENSMUST00000217480.2
ENSMUST00000215409.2 |
Olfr54
|
olfactory receptor 54 |
| chr17_-_22792463 | 0.54 |
ENSMUST00000092491.7
ENSMUST00000234223.2 ENSMUST00000234296.2 ENSMUST00000234027.2 |
Vmn2r111
|
vomeronasal 2, receptor 111 |
| chr5_-_135494775 | 0.45 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr6_-_146403638 | 0.45 |
ENSMUST00000079573.13
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr17_-_38370970 | 0.42 |
ENSMUST00000216476.2
|
Olfr129
|
olfactory receptor 129 |
| chr11_-_71092282 | 0.36 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr6_+_136495818 | 0.35 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr11_+_96822213 | 0.28 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr16_-_95993420 | 0.26 |
ENSMUST00000113804.8
ENSMUST00000054855.14 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chr11_-_71092124 | 0.23 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr10_-_14593935 | 0.21 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr2_+_172314433 | 0.21 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr6_+_136495784 | 0.18 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_106630381 | 0.17 |
ENSMUST00000213623.2
|
Olfr713
|
olfactory receptor 713 |
| chr16_+_31247562 | 0.13 |
ENSMUST00000115227.10
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr6_+_125297596 | 0.11 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr11_-_75345482 | 0.11 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chr10_-_117118226 | 0.09 |
ENSMUST00000092163.9
|
Lyz2
|
lysozyme 2 |
| chr3_-_95811993 | 0.07 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chr17_+_23562715 | 0.06 |
ENSMUST00000168175.3
ENSMUST00000234796.2 |
Vmn2r115
|
vomeronasal 2, receptor 115 |
| chr15_+_102011352 | 0.05 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr8_-_95328348 | 0.04 |
ENSMUST00000212547.2
ENSMUST00000212507.2 ENSMUST00000034226.8 |
Psme3ip1
|
proteasome activator subunit 3 interacting protein 1 |
| chr5_+_31855009 | 0.03 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr15_+_102011415 | 0.01 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr5_+_146418775 | 0.01 |
ENSMUST00000179032.3
|
Gm6408
|
predicted gene 6408 |
| chr14_+_44822935 | 0.01 |
ENSMUST00000178759.2
|
Gm8247
|
predicted gene 8247 |
| chr6_-_69394425 | 0.01 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr6_-_69037208 | 0.01 |
ENSMUST00000103343.4
|
Igkv4-78
|
immunoglobulin kappa variable 4-78 |
| chr6_-_69245427 | 0.00 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr2_-_164435508 | 0.00 |
ENSMUST00000103100.2
|
Eppin
|
epididymal peptidase inhibitor |
| chr5_-_146097347 | 0.00 |
ENSMUST00000199463.2
|
Gm5565
|
predicted gene 5565 |
| chr4_-_49408040 | 0.00 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr5_-_146107531 | 0.00 |
ENSMUST00000174320.2
|
Gm6309
|
predicted gene 6309 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr3c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.2 | 1.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.6 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.1 | 0.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 1.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.6 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 1.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |