Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nr4a2
Z-value: 0.41
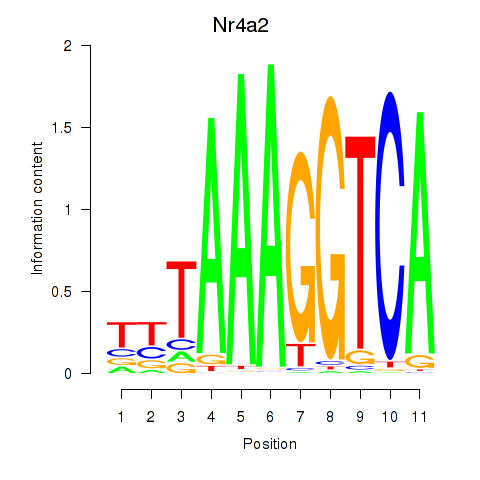
Transcription factors associated with Nr4a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a2
|
ENSMUSG00000026826.14 | nuclear receptor subfamily 4, group A, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a2 | mm39_v1_chr2_-_57004933_57005050 | 0.30 | 6.2e-01 | Click! |
Activity profile of Nr4a2 motif
Sorted Z-values of Nr4a2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_35587888 | 0.29 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr10_-_40178182 | 0.25 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr4_+_99812912 | 0.24 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr4_+_102617495 | 0.24 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr4_+_102617332 | 0.21 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr6_-_124791259 | 0.18 |
ENSMUST00000172132.10
ENSMUST00000239432.2 |
Tpi1
|
triosephosphate isomerase 1 |
| chr11_+_70548022 | 0.17 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr7_+_120234399 | 0.16 |
ENSMUST00000033176.7
ENSMUST00000208400.2 |
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr2_-_73741664 | 0.16 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr7_-_100164007 | 0.15 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr8_-_65186565 | 0.15 |
ENSMUST00000141021.2
|
Msmo1
|
methylsterol monoxygenase 1 |
| chr1_-_63215812 | 0.14 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr13_-_98152768 | 0.13 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr8_-_46664321 | 0.13 |
ENSMUST00000034049.5
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr14_-_52252003 | 0.13 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chrX_+_7445806 | 0.12 |
ENSMUST00000234363.2
ENSMUST00000235116.2 ENSMUST00000115739.9 ENSMUST00000234574.2 ENSMUST00000115740.9 |
Foxp3
|
forkhead box P3 |
| chrX_-_72965434 | 0.12 |
ENSMUST00000096316.4
ENSMUST00000114390.8 ENSMUST00000114391.10 ENSMUST00000114387.8 |
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr4_+_97665843 | 0.11 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr15_+_102381929 | 0.11 |
ENSMUST00000229746.2
ENSMUST00000230211.2 ENSMUST00000230539.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr5_+_135216090 | 0.11 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr2_+_128907854 | 0.11 |
ENSMUST00000035812.14
|
Ttl
|
tubulin tyrosine ligase |
| chr19_-_32038838 | 0.10 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr11_-_109363406 | 0.10 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr18_-_57108405 | 0.10 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr1_-_63215952 | 0.10 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr2_-_101451383 | 0.09 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chrX_+_149372903 | 0.09 |
ENSMUST00000080884.11
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_-_98505508 | 0.09 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr2_-_25911544 | 0.09 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr16_-_20245138 | 0.08 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr10_+_126914755 | 0.08 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_+_43960183 | 0.08 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_-_163259012 | 0.08 |
ENSMUST00000127038.2
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr3_+_84832783 | 0.08 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr3_+_89344006 | 0.08 |
ENSMUST00000038942.10
ENSMUST00000130858.8 ENSMUST00000146630.8 ENSMUST00000145753.2 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr18_+_36797113 | 0.08 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr5_-_90487583 | 0.08 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chrX_-_47602395 | 0.08 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr15_+_102381705 | 0.07 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr11_+_70548622 | 0.07 |
ENSMUST00000170716.8
|
Eno3
|
enolase 3, beta muscle |
| chr19_+_43770619 | 0.07 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr4_-_150998857 | 0.07 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr14_-_52252432 | 0.07 |
ENSMUST00000226527.2
|
Zfp219
|
zinc finger protein 219 |
| chr15_-_102425241 | 0.07 |
ENSMUST00000169162.8
ENSMUST00000023812.10 ENSMUST00000165174.8 ENSMUST00000169367.8 ENSMUST00000169377.8 |
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chrX_-_72965524 | 0.07 |
ENSMUST00000114389.10
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr5_-_88823472 | 0.07 |
ENSMUST00000113234.8
ENSMUST00000153565.8 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chrX_-_72965536 | 0.07 |
ENSMUST00000033763.15
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr2_-_165125519 | 0.06 |
ENSMUST00000155289.8
|
Slc35c2
|
solute carrier family 35, member C2 |
| chr2_-_25911691 | 0.06 |
ENSMUST00000036509.14
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr1_-_190950129 | 0.05 |
ENSMUST00000195117.2
|
Atf3
|
activating transcription factor 3 |
| chr7_+_51537645 | 0.05 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr2_+_58991182 | 0.05 |
ENSMUST00000168631.8
ENSMUST00000102754.11 ENSMUST00000123908.8 |
Pkp4
|
plakophilin 4 |
| chr5_-_88823049 | 0.05 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chrX_-_139501246 | 0.05 |
ENSMUST00000112996.9
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr19_+_55240357 | 0.05 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr14_-_52252318 | 0.05 |
ENSMUST00000228051.2
|
Zfp219
|
zinc finger protein 219 |
| chr2_+_158636727 | 0.04 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr10_-_99595498 | 0.04 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr1_+_63216281 | 0.04 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr6_-_125213911 | 0.04 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chrX_+_151016224 | 0.04 |
ENSMUST00000112588.9
ENSMUST00000082177.13 |
Kdm5c
|
lysine (K)-specific demethylase 5C |
| chr2_+_75662806 | 0.04 |
ENSMUST00000175646.2
|
Agps
|
alkylglycerone phosphate synthase |
| chr15_+_76579960 | 0.03 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr9_-_42372710 | 0.03 |
ENSMUST00000066179.14
|
Tbcel
|
tubulin folding cofactor E-like |
| chr7_+_24310171 | 0.03 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr2_+_75662511 | 0.03 |
ENSMUST00000047232.14
ENSMUST00000111952.9 |
Agps
|
alkylglycerone phosphate synthase |
| chrX_-_141173330 | 0.03 |
ENSMUST00000112907.8
|
Acsl4
|
acyl-CoA synthetase long-chain family member 4 |
| chr16_-_43959559 | 0.03 |
ENSMUST00000063661.13
ENSMUST00000114666.9 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr13_+_93440265 | 0.03 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chr10_-_42354276 | 0.03 |
ENSMUST00000151747.8
|
Afg1l
|
AFG1 like ATPase |
| chr9_-_21996693 | 0.03 |
ENSMUST00000179422.8
ENSMUST00000098937.10 ENSMUST00000177967.2 ENSMUST00000180180.8 |
Ecsit
|
ECSIT signalling integrator |
| chr5_+_30437579 | 0.03 |
ENSMUST00000145167.9
|
Selenoi
|
selenoprotein I |
| chr10_+_67021509 | 0.03 |
ENSMUST00000173689.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr19_-_5168251 | 0.02 |
ENSMUST00000113728.8
ENSMUST00000113727.8 ENSMUST00000025798.13 |
Klc2
|
kinesin light chain 2 |
| chr5_-_121665249 | 0.02 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr4_+_97665992 | 0.02 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr12_-_75596441 | 0.02 |
ENSMUST00000218716.2
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr4_+_41966058 | 0.02 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr19_+_3817396 | 0.02 |
ENSMUST00000052699.13
ENSMUST00000113974.11 ENSMUST00000113972.9 ENSMUST00000113973.8 ENSMUST00000113977.9 ENSMUST00000113968.9 |
Kmt5b
|
lysine methyltransferase 5B |
| chr5_-_31359276 | 0.02 |
ENSMUST00000031562.11
|
Zfp513
|
zinc finger protein 513 |
| chr9_+_118335294 | 0.02 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr5_-_31359559 | 0.02 |
ENSMUST00000202929.2
ENSMUST00000201231.2 ENSMUST00000114590.8 |
Zfp513
|
zinc finger protein 513 |
| chr6_+_92068361 | 0.02 |
ENSMUST00000113460.8
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr10_-_42354482 | 0.02 |
ENSMUST00000041024.15
|
Afg1l
|
AFG1 like ATPase |
| chr4_-_42853888 | 0.02 |
ENSMUST00000107979.2
|
Fam205a1
|
family with sequence similarity 205, member A1 |
| chr6_+_113674011 | 0.02 |
ENSMUST00000089018.11
|
Tatdn2
|
TatD DNase domain containing 2 |
| chr9_+_55056648 | 0.02 |
ENSMUST00000121677.8
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr13_+_93440572 | 0.02 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr5_+_150183201 | 0.02 |
ENSMUST00000087204.9
|
Fry
|
FRY microtubule binding protein |
| chr10_+_26105605 | 0.02 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr4_+_42318272 | 0.01 |
ENSMUST00000178192.3
|
Fam205a3
|
family with sequence similarity 205, member A3 |
| chr16_+_8647959 | 0.01 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr16_-_20245071 | 0.01 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr9_+_108270020 | 0.01 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr15_+_25774070 | 0.01 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr11_+_96214078 | 0.01 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr10_+_110756031 | 0.01 |
ENSMUST00000220409.2
ENSMUST00000219502.2 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr11_+_70548513 | 0.01 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr1_+_63215976 | 0.01 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chrX_-_149372840 | 0.01 |
ENSMUST00000112725.8
ENSMUST00000112720.8 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr11_+_121593582 | 0.01 |
ENSMUST00000125580.2
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr16_+_21644692 | 0.01 |
ENSMUST00000232240.2
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr12_-_76842263 | 0.01 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr18_+_77861656 | 0.01 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr9_-_70842090 | 0.00 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr19_+_36532061 | 0.00 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr5_+_88523967 | 0.00 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr9_+_108269992 | 0.00 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr6_+_40470463 | 0.00 |
ENSMUST00000038750.7
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr14_-_78326435 | 0.00 |
ENSMUST00000118785.3
ENSMUST00000066437.5 |
Fam216b
|
family with sequence similarity 216, member B |
| chr15_+_102875229 | 0.00 |
ENSMUST00000001699.8
|
Hoxc10
|
homeobox C10 |
| chr6_-_50631418 | 0.00 |
ENSMUST00000031853.8
|
Npvf
|
neuropeptide VF precursor |
| chr11_-_24025054 | 0.00 |
ENSMUST00000068360.2
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr11_-_69838971 | 0.00 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr16_-_58850641 | 0.00 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chr8_-_13544478 | 0.00 |
ENSMUST00000033828.7
|
Gas6
|
growth arrest specific 6 |
| chr15_+_25940912 | 0.00 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr10_-_41179167 | 0.00 |
ENSMUST00000043814.5
|
Fig4
|
FIG4 phosphoinositide 5-phosphatase |
| chr4_-_36136463 | 0.00 |
ENSMUST00000098151.9
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chrX_-_166638057 | 0.00 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr11_-_109613040 | 0.00 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr16_-_19443851 | 0.00 |
ENSMUST00000079891.4
|
Olfr171
|
olfactory receptor 171 |
| chr14_-_8798841 | 0.00 |
ENSMUST00000061045.3
|
Sntn
|
sentan, cilia apical structure protein |
| chr14_+_27598021 | 0.00 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr5_-_67256578 | 0.00 |
ENSMUST00000012664.11
|
Phox2b
|
paired-like homeobox 2b |
| chr9_-_106533279 | 0.00 |
ENSMUST00000023959.13
ENSMUST00000201681.2 |
Grm2
|
glutamate receptor, metabotropic 2 |
| chr6_-_138399896 | 0.00 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr9_-_70841881 | 0.00 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr10_+_27950809 | 0.00 |
ENSMUST00000166468.2
ENSMUST00000218359.2 ENSMUST00000218276.2 |
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.1 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.2 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0002849 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.2 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.1 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |