Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Nr6a1
Z-value: 0.76
Transcription factors associated with Nr6a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr6a1
|
ENSMUSG00000063972.14 | nuclear receptor subfamily 6, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr6a1 | mm39_v1_chr2_-_38816229_38816366 | 0.68 | 2.0e-01 | Click! |
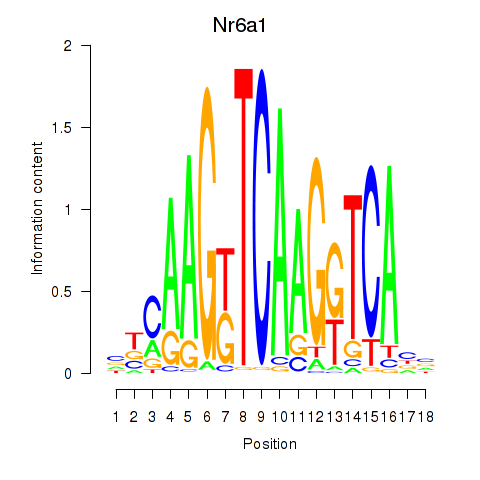
Activity profile of Nr6a1 motif
Sorted Z-values of Nr6a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_64969679 | 0.61 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr9_+_87026337 | 0.57 |
ENSMUST00000113149.8
ENSMUST00000049457.14 ENSMUST00000179313.3 |
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr7_+_110368037 | 0.56 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr12_+_44268134 | 0.55 |
ENSMUST00000122902.8
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr15_+_84553801 | 0.44 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr16_+_11140740 | 0.38 |
ENSMUST00000180792.8
|
Snx29
|
sorting nexin 29 |
| chr18_-_6135888 | 0.38 |
ENSMUST00000182383.8
ENSMUST00000062584.14 ENSMUST00000077128.13 ENSMUST00000182038.2 ENSMUST00000182213.8 ENSMUST00000182559.8 |
Arhgap12
|
Rho GTPase activating protein 12 |
| chr12_-_115916604 | 0.37 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr12_-_115832846 | 0.36 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr5_-_108822619 | 0.35 |
ENSMUST00000119270.2
ENSMUST00000163328.8 ENSMUST00000136227.2 |
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr14_-_121742659 | 0.34 |
ENSMUST00000088386.8
|
Slc15a1
|
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr16_+_11140779 | 0.33 |
ENSMUST00000115814.4
|
Snx29
|
sorting nexin 29 |
| chr5_-_137609691 | 0.32 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr12_-_114547622 | 0.27 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chr2_+_127267069 | 0.27 |
ENSMUST00000062211.4
|
Gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr4_+_123798690 | 0.26 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr6_+_21985902 | 0.26 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_137609634 | 0.24 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr15_-_77653979 | 0.24 |
ENSMUST00000229259.2
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr15_+_99936516 | 0.19 |
ENSMUST00000100203.10
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr5_-_36641456 | 0.17 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr12_-_115884332 | 0.17 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr17_+_33848054 | 0.16 |
ENSMUST00000166627.8
ENSMUST00000073570.12 ENSMUST00000170225.3 |
Zfp414
|
zinc finger protein 414 |
| chr12_-_114878652 | 0.15 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr12_-_114752425 | 0.15 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr5_-_5744559 | 0.14 |
ENSMUST00000115426.9
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr15_-_50753061 | 0.14 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr12_-_115825934 | 0.13 |
ENSMUST00000198777.2
|
Ighv1-77
|
immunoglobulin heavy variable 1-77 |
| chr12_-_114672701 | 0.12 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr13_+_113931038 | 0.12 |
ENSMUST00000091201.7
|
Arl15
|
ADP-ribosylation factor-like 15 |
| chr5_-_5744024 | 0.12 |
ENSMUST00000115425.9
ENSMUST00000115427.8 ENSMUST00000115424.9 ENSMUST00000015797.11 |
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr12_-_114646685 | 0.12 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr5_-_137608886 | 0.11 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr2_+_172314433 | 0.11 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr18_+_65020910 | 0.10 |
ENSMUST00000236736.2
ENSMUST00000235743.2 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr18_+_65020760 | 0.10 |
ENSMUST00000235343.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr18_+_65020885 | 0.10 |
ENSMUST00000236209.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr17_+_73225292 | 0.10 |
ENSMUST00000024857.14
|
Lbh
|
limb-bud and heart |
| chr1_+_151304293 | 0.09 |
ENSMUST00000065625.12
|
Trmt1l
|
tRNA methyltransferase 1 like |
| chr5_-_137015683 | 0.09 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr6_-_67468831 | 0.09 |
ENSMUST00000118364.2
|
Il23r
|
interleukin 23 receptor |
| chr16_-_3535958 | 0.09 |
ENSMUST00000023180.15
|
Mefv
|
Mediterranean fever |
| chr12_-_115299134 | 0.09 |
ENSMUST00000195359.6
ENSMUST00000103530.3 |
Ighv1-59
|
immunoglobulin heavy variable V1-59 |
| chr9_+_50515207 | 0.08 |
ENSMUST00000044051.6
|
Timm8b
|
translocase of inner mitochondrial membrane 8B |
| chr16_-_3535940 | 0.08 |
ENSMUST00000229725.2
ENSMUST00000100222.4 |
Mefv
|
Mediterranean fever |
| chr12_-_114621406 | 0.08 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr9_+_122634589 | 0.08 |
ENSMUST00000052740.14
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr5_-_143278619 | 0.06 |
ENSMUST00000212355.2
|
Zfp853
|
zinc finger protein 853 |
| chr18_+_65158873 | 0.06 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr12_-_114843941 | 0.06 |
ENSMUST00000191862.6
ENSMUST00000103513.3 |
Ighv1-36
|
immunoglobulin heavy variable 1-36 |
| chr19_-_5738177 | 0.06 |
ENSMUST00000068169.12
|
Pcnx3
|
pecanex homolog 3 |
| chr9_+_38382403 | 0.06 |
ENSMUST00000214377.2
|
Olfr905
|
olfactory receptor 905 |
| chr5_+_129802127 | 0.05 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr10_+_126911134 | 0.05 |
ENSMUST00000239120.2
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr3_+_107137924 | 0.05 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr7_+_133239272 | 0.05 |
ENSMUST00000051169.13
|
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr5_-_123711098 | 0.05 |
ENSMUST00000031388.13
|
Vps33a
|
VPS33A CORVET/HOPS core subunit |
| chr1_-_79836344 | 0.04 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr4_-_45530330 | 0.04 |
ENSMUST00000061986.12
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr5_+_137015873 | 0.04 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr1_-_151304191 | 0.04 |
ENSMUST00000064771.12
|
Swt1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr1_+_82702598 | 0.03 |
ENSMUST00000078332.13
ENSMUST00000073025.12 ENSMUST00000161648.8 ENSMUST00000160786.8 ENSMUST00000162003.8 |
Mff
|
mitochondrial fission factor |
| chr1_+_160806194 | 0.03 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr4_-_124959395 | 0.03 |
ENSMUST00000036383.4
|
Dnali1
|
dynein, axonemal, light intermediate polypeptide 1 |
| chr4_-_138858340 | 0.03 |
ENSMUST00000143971.2
|
Micos10
|
mitochondrial contact site and cristae organizing system subunit 10 |
| chr12_-_113896002 | 0.03 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr13_-_25204272 | 0.03 |
ENSMUST00000021772.4
|
Mrs2
|
MRS2 magnesium transporter |
| chr7_+_127864847 | 0.03 |
ENSMUST00000118169.8
ENSMUST00000206909.2 ENSMUST00000142841.8 |
Slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr4_+_123798625 | 0.03 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr11_-_100661762 | 0.02 |
ENSMUST00000139341.2
ENSMUST00000017891.14 |
Ghdc
|
GH3 domain containing |
| chr16_+_9988080 | 0.02 |
ENSMUST00000121292.8
ENSMUST00000044103.6 |
Rpl39l
|
ribosomal protein L39-like |
| chr15_+_27466732 | 0.02 |
ENSMUST00000022875.7
|
Ank
|
progressive ankylosis |
| chr12_-_114815280 | 0.02 |
ENSMUST00000103512.3
|
Ighv1-34
|
immunoglobulin heavy variable 1-34 |
| chr19_+_13037974 | 0.02 |
ENSMUST00000213806.2
ENSMUST00000214695.2 ENSMUST00000217568.2 |
Olfr1454
|
olfactory receptor 1454 |
| chr5_+_36641922 | 0.01 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr12_-_11200306 | 0.01 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr3_-_152232389 | 0.01 |
ENSMUST00000200062.2
|
Ak5
|
adenylate kinase 5 |
| chr9_-_121668527 | 0.01 |
ENSMUST00000135986.9
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr4_-_82423511 | 0.01 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chr11_-_69712970 | 0.01 |
ENSMUST00000045771.7
|
Spem1
|
sperm maturation 1 |
| chr5_+_30437579 | 0.01 |
ENSMUST00000145167.9
|
Selenoi
|
selenoprotein I |
| chr9_+_45311000 | 0.00 |
ENSMUST00000216289.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr7_+_44890497 | 0.00 |
ENSMUST00000213347.2
|
Slc6a16
|
solute carrier family 6, member 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr6a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.2 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 1.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.6 | GO:0097009 | energy homeostasis(GO:0097009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.3 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 1.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |