Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Obox1
Z-value: 1.23
Transcription factors associated with Obox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Obox1
|
ENSMUSG00000054310.17 | oocyte specific homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox1 | mm39_v1_chr7_+_15246265_15246265 | 0.40 | 5.0e-01 | Click! |
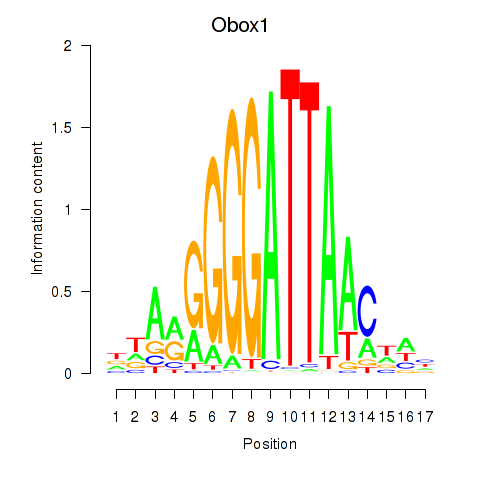
Activity profile of Obox1 motif
Sorted Z-values of Obox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_142202642 | 0.75 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr13_+_55862437 | 0.74 |
ENSMUST00000021959.11
|
Txndc15
|
thioredoxin domain containing 15 |
| chr11_-_68864666 | 0.70 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr11_+_59738866 | 0.67 |
ENSMUST00000102695.4
|
Nt5m
|
5',3'-nucleotidase, mitochondrial |
| chr19_-_33567708 | 0.56 |
ENSMUST00000112508.9
|
Lipo3
|
lipase, member O3 |
| chr12_-_101943134 | 0.55 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr1_-_135241429 | 0.54 |
ENSMUST00000134088.3
ENSMUST00000081104.10 |
Timm17a
|
translocase of inner mitochondrial membrane 17a |
| chr12_-_36206780 | 0.54 |
ENSMUST00000223382.2
ENSMUST00000020856.6 |
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_-_11618192 | 0.54 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr19_-_11618165 | 0.51 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr17_+_24937062 | 0.50 |
ENSMUST00000152407.8
|
Rps2
|
ribosomal protein S2 |
| chr11_+_109540201 | 0.50 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr16_+_48692976 | 0.50 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr6_+_58810674 | 0.48 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr2_-_126342551 | 0.46 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr17_+_21876498 | 0.45 |
ENSMUST00000039726.8
|
Zfp983
|
zinc finger protein 983 |
| chr11_-_62172164 | 0.44 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr6_-_113717689 | 0.44 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr3_+_142406787 | 0.43 |
ENSMUST00000106218.8
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr11_-_116164928 | 0.43 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr14_-_31362835 | 0.43 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr3_+_142406827 | 0.43 |
ENSMUST00000044392.11
ENSMUST00000199519.5 |
Kyat3
|
kynurenine aminotransferase 3 |
| chr7_+_46496929 | 0.43 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr6_-_129599645 | 0.42 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr9_-_106768601 | 0.42 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr11_-_88742285 | 0.41 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr19_-_10079091 | 0.41 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr18_+_24122689 | 0.40 |
ENSMUST00000074941.8
|
Zfp35
|
zinc finger protein 35 |
| chr11_-_82799186 | 0.40 |
ENSMUST00000103213.10
|
Nle1
|
notchless homolog 1 |
| chr10_-_128640232 | 0.40 |
ENSMUST00000051011.14
|
Tmem198b
|
transmembrane protein 198b |
| chr7_+_142606476 | 0.40 |
ENSMUST00000037941.10
|
Cd81
|
CD81 antigen |
| chr3_+_32760447 | 0.40 |
ENSMUST00000194781.6
|
Actl6a
|
actin-like 6A |
| chr14_+_31888061 | 0.40 |
ENSMUST00000164341.2
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr15_+_76227695 | 0.39 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr2_+_19376447 | 0.39 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr2_+_144435974 | 0.39 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr12_-_13299197 | 0.38 |
ENSMUST00000071103.10
|
Ddx1
|
DEAD box helicase 1 |
| chr15_+_72962241 | 0.37 |
ENSMUST00000089765.9
ENSMUST00000134353.2 |
Chrac1
|
chromatin accessibility complex 1 |
| chr1_-_60137294 | 0.37 |
ENSMUST00000141417.3
ENSMUST00000122038.8 |
Wdr12
|
WD repeat domain 12 |
| chr7_+_18883647 | 0.36 |
ENSMUST00000049294.4
|
Snrpd2
|
small nuclear ribonucleoprotein D2 |
| chr11_+_95275458 | 0.36 |
ENSMUST00000021243.16
ENSMUST00000146556.2 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr11_+_95925711 | 0.36 |
ENSMUST00000006217.10
ENSMUST00000107700.4 |
Snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr7_-_24287037 | 0.36 |
ENSMUST00000094705.3
|
Zfp575
|
zinc finger protein 575 |
| chr8_-_4829519 | 0.36 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr11_+_29668563 | 0.36 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr11_-_53321242 | 0.35 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr18_+_67338437 | 0.34 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr7_+_16186704 | 0.34 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chrX_+_158155171 | 0.34 |
ENSMUST00000087143.7
|
Eif1ax
|
eukaryotic translation initiation factor 1A, X-linked |
| chr10_+_61531282 | 0.34 |
ENSMUST00000020284.5
|
Tysnd1
|
trypsin domain containing 1 |
| chr1_-_10108325 | 0.33 |
ENSMUST00000027050.10
ENSMUST00000188619.2 |
Cops5
|
COP9 signalosome subunit 5 |
| chr14_+_55129950 | 0.33 |
ENSMUST00000140691.8
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr10_-_75616788 | 0.33 |
ENSMUST00000173512.2
ENSMUST00000173537.2 |
Gm20441
Gstt3
|
predicted gene 20441 glutathione S-transferase, theta 3 |
| chr16_+_33614378 | 0.32 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chrX_-_101232978 | 0.32 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr19_-_10859046 | 0.32 |
ENSMUST00000128835.8
|
Tmem109
|
transmembrane protein 109 |
| chr19_+_32573182 | 0.31 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr10_+_79977291 | 0.31 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chrX_+_133618693 | 0.31 |
ENSMUST00000113201.8
ENSMUST00000051256.10 ENSMUST00000113199.8 ENSMUST00000035748.14 ENSMUST00000113198.8 ENSMUST00000113197.2 |
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr11_-_116165024 | 0.31 |
ENSMUST00000021133.16
|
Srp68
|
signal recognition particle 68 |
| chr19_+_24853039 | 0.31 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr1_-_60137263 | 0.31 |
ENSMUST00000143342.8
|
Wdr12
|
WD repeat domain 12 |
| chr7_+_81412695 | 0.31 |
ENSMUST00000133034.2
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr9_+_73020708 | 0.30 |
ENSMUST00000169399.8
ENSMUST00000034738.14 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chr11_-_120687195 | 0.30 |
ENSMUST00000143139.8
ENSMUST00000129955.2 ENSMUST00000026151.11 ENSMUST00000167023.8 ENSMUST00000106133.8 ENSMUST00000106135.8 |
Dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr11_-_86148379 | 0.29 |
ENSMUST00000132024.8
ENSMUST00000139285.8 |
Ints2
|
integrator complex subunit 2 |
| chr17_-_26016039 | 0.29 |
ENSMUST00000165838.9
ENSMUST00000002344.7 |
Metrn
|
meteorin, glial cell differentiation regulator |
| chr3_-_108053396 | 0.29 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr11_-_69470139 | 0.29 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr13_+_41169740 | 0.29 |
ENSMUST00000021790.7
|
Tmem14c
|
transmembrane protein 14C |
| chr12_-_108859123 | 0.29 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr7_-_113875261 | 0.28 |
ENSMUST00000135570.8
|
Psma1
|
proteasome subunit alpha 1 |
| chr5_-_137530214 | 0.28 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_-_153425791 | 0.28 |
ENSMUST00000041874.9
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr18_-_66155651 | 0.27 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr4_-_75196485 | 0.27 |
ENSMUST00000030103.9
|
Dmac1
|
distal membrane arm assembly complex 1 |
| chr13_-_99481160 | 0.27 |
ENSMUST00000022153.8
|
Ptcd2
|
pentatricopeptide repeat domain 2 |
| chr9_-_57375269 | 0.27 |
ENSMUST00000215059.2
ENSMUST00000046587.8 ENSMUST00000214256.2 |
Scamp5
|
secretory carrier membrane protein 5 |
| chr4_+_119052548 | 0.27 |
ENSMUST00000106345.3
|
Svbp
|
small vasohibin binding protein |
| chr7_+_133311062 | 0.27 |
ENSMUST00000033282.5
|
Bccip
|
BRCA2 and CDKN1A interacting protein |
| chr2_-_23939401 | 0.27 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr7_+_81412673 | 0.27 |
ENSMUST00000042166.11
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr4_+_116876597 | 0.26 |
ENSMUST00000106448.9
ENSMUST00000106447.9 |
Eif2b3
|
eukaryotic translation initiation factor 2B, subunit 3 |
| chr13_-_54616618 | 0.26 |
ENSMUST00000026990.6
|
Thoc3
|
THO complex 3 |
| chr16_-_30900181 | 0.26 |
ENSMUST00000055389.9
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr9_-_53521585 | 0.26 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr1_-_53391778 | 0.25 |
ENSMUST00000236737.2
ENSMUST00000027264.10 ENSMUST00000123519.9 |
Gm50478
Asnsd1
|
predicted gene, 50478 asparagine synthetase domain containing 1 |
| chr4_+_101504938 | 0.25 |
ENSMUST00000106927.2
|
Leprot
|
leptin receptor overlapping transcript |
| chr14_+_52348396 | 0.25 |
ENSMUST00000111600.11
|
Rpgrip1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr16_-_91525655 | 0.25 |
ENSMUST00000117644.8
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr5_+_21942139 | 0.25 |
ENSMUST00000030882.12
|
Pmpcb
|
peptidase (mitochondrial processing) beta |
| chr15_+_81548090 | 0.25 |
ENSMUST00000023029.15
ENSMUST00000174229.8 ENSMUST00000172748.8 |
L3mbtl2
|
L3MBTL2 polycomb repressive complex 1 subunit |
| chr16_+_31241085 | 0.24 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr7_-_44635740 | 0.24 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr5_-_134643805 | 0.24 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr5_-_65248927 | 0.23 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr1_+_163889713 | 0.22 |
ENSMUST00000097491.10
|
Sell
|
selectin, lymphocyte |
| chr16_+_33614715 | 0.22 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr11_+_3913970 | 0.22 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chrY_+_1010543 | 0.21 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr14_-_70043079 | 0.21 |
ENSMUST00000022665.4
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chrX_+_141010919 | 0.21 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_181506130 | 0.21 |
ENSMUST00000039551.9
|
Polr3k
|
polymerase (RNA) III (DNA directed) polypeptide K |
| chr15_+_100659622 | 0.20 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr3_-_63872189 | 0.20 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr15_+_97682210 | 0.20 |
ENSMUST00000117892.2
ENSMUST00000229084.2 |
Slc48a1
|
solute carrier family 48 (heme transporter), member 1 |
| chr15_-_83006957 | 0.20 |
ENSMUST00000018184.10
|
Rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr7_+_37883300 | 0.19 |
ENSMUST00000179992.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr13_-_98628509 | 0.19 |
ENSMUST00000170205.2
|
Gm10320
|
predicted pseudogene 10320 |
| chr18_-_32044877 | 0.19 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr8_+_114860375 | 0.19 |
ENSMUST00000147605.8
ENSMUST00000134593.2 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr16_-_91525863 | 0.19 |
ENSMUST00000073466.13
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr11_+_115044966 | 0.19 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr15_-_76544308 | 0.19 |
ENSMUST00000066677.10
ENSMUST00000177359.2 |
Cyhr1
|
cysteine and histidine rich 1 |
| chr4_+_155789246 | 0.19 |
ENSMUST00000030905.9
|
Ssu72
|
Ssu72 RNA polymerase II CTD phosphatase homolog (yeast) |
| chr9_-_21223551 | 0.19 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr1_-_52271455 | 0.18 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr5_-_137529465 | 0.18 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_-_113875342 | 0.18 |
ENSMUST00000033008.10
|
Psma1
|
proteasome subunit alpha 1 |
| chr17_-_35265702 | 0.18 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr17_-_35984625 | 0.18 |
ENSMUST00000001565.15
|
Gtf2h4
|
general transcription factor II H, polypeptide 4 |
| chr2_-_86109346 | 0.17 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr16_+_55786638 | 0.17 |
ENSMUST00000023269.5
|
Rpl24
|
ribosomal protein L24 |
| chr9_+_62249730 | 0.17 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr17_+_26780453 | 0.17 |
ENSMUST00000167662.8
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr18_+_31922173 | 0.17 |
ENSMUST00000025106.5
ENSMUST00000234146.2 |
Polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr10_-_117582259 | 0.17 |
ENSMUST00000079041.7
|
Slc35e3
|
solute carrier family 35, member E3 |
| chr11_-_30599510 | 0.17 |
ENSMUST00000074613.4
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr6_-_69282389 | 0.17 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr2_-_28511941 | 0.16 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr14_-_20027112 | 0.16 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr2_-_152185901 | 0.16 |
ENSMUST00000040312.7
|
Trib3
|
tribbles pseudokinase 3 |
| chr11_+_69886652 | 0.16 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr19_-_6919755 | 0.16 |
ENSMUST00000099782.10
|
Gpr137
|
G protein-coupled receptor 137 |
| chr7_+_88079465 | 0.15 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr16_-_17906886 | 0.15 |
ENSMUST00000132241.2
ENSMUST00000139861.2 ENSMUST00000003620.13 |
Prodh
|
proline dehydrogenase |
| chrX_-_52645649 | 0.15 |
ENSMUST00000088779.5
|
Rtl8a
|
retrotransposon Gag like 8A |
| chr5_+_124690908 | 0.15 |
ENSMUST00000071057.14
ENSMUST00000111438.2 |
Ddx55
|
DEAD box helicase 55 |
| chr7_+_101871623 | 0.15 |
ENSMUST00000143541.8
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr11_+_4852212 | 0.15 |
ENSMUST00000142543.3
|
Thoc5
|
THO complex 5 |
| chr9_+_62249341 | 0.14 |
ENSMUST00000135395.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr16_+_35861554 | 0.14 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr14_+_59438658 | 0.14 |
ENSMUST00000173547.8
ENSMUST00000043227.13 ENSMUST00000022551.14 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr11_+_23616007 | 0.14 |
ENSMUST00000058163.11
|
Pus10
|
pseudouridylate synthase 10 |
| chr11_-_59937302 | 0.14 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr1_+_134487893 | 0.14 |
ENSMUST00000047714.14
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr15_+_76579960 | 0.14 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr3_+_151143557 | 0.14 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr17_-_12726591 | 0.14 |
ENSMUST00000024595.4
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr19_-_6178171 | 0.14 |
ENSMUST00000154601.8
ENSMUST00000138931.3 |
Snx15
|
sorting nexin 15 |
| chr8_+_79236051 | 0.14 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr7_-_44635813 | 0.13 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr11_+_6242555 | 0.13 |
ENSMUST00000081894.5
|
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr4_-_116982804 | 0.13 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr8_-_84184978 | 0.13 |
ENSMUST00000081506.11
|
Scoc
|
short coiled-coil protein |
| chr5_-_65248962 | 0.13 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr11_-_86148344 | 0.13 |
ENSMUST00000136469.2
ENSMUST00000018212.13 |
Ints2
|
integrator complex subunit 2 |
| chr12_-_102390000 | 0.13 |
ENSMUST00000110020.8
|
Lgmn
|
legumain |
| chr2_-_34803988 | 0.13 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr17_+_36132567 | 0.13 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr2_+_83786037 | 0.13 |
ENSMUST00000179192.2
|
Gm13698
|
predicted gene 13698 |
| chr11_+_23616477 | 0.13 |
ENSMUST00000143117.2
|
Pus10
|
pseudouridylate synthase 10 |
| chr1_-_139708906 | 0.12 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr3_-_63872079 | 0.12 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr16_-_18161746 | 0.11 |
ENSMUST00000231372.2
ENSMUST00000130752.2 ENSMUST00000231605.2 ENSMUST00000115628.10 |
Tango2
|
transport and golgi organization 2 |
| chr13_+_58550499 | 0.11 |
ENSMUST00000225815.2
|
Rmi1
|
RecQ mediated genome instability 1 |
| chr11_-_5657658 | 0.11 |
ENSMUST00000154330.2
|
Mrps24
|
mitochondrial ribosomal protein S24 |
| chr11_-_79145489 | 0.11 |
ENSMUST00000017821.12
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr10_-_79875479 | 0.11 |
ENSMUST00000004786.10
|
Polr2e
|
polymerase (RNA) II (DNA directed) polypeptide E |
| chr10_+_94034817 | 0.11 |
ENSMUST00000020209.16
ENSMUST00000179990.8 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chrX_-_84820250 | 0.11 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr5_+_143450329 | 0.11 |
ENSMUST00000045593.12
|
Daglb
|
diacylglycerol lipase, beta |
| chr11_-_102120917 | 0.10 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr6_-_124710084 | 0.10 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_+_124908341 | 0.10 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr2_-_87902174 | 0.10 |
ENSMUST00000215268.2
|
Olfr1163
|
olfactory receptor 1163 |
| chr7_+_27198740 | 0.10 |
ENSMUST00000098644.9
ENSMUST00000108355.2 ENSMUST00000238936.2 |
Prx
|
periaxin |
| chr17_+_66418525 | 0.10 |
ENSMUST00000072383.14
|
Washc1
|
WASH complex subunit 1 |
| chr12_+_86725459 | 0.10 |
ENSMUST00000021681.4
|
Vash1
|
vasohibin 1 |
| chr1_+_173248104 | 0.10 |
ENSMUST00000173023.2
|
Aim2
|
absent in melanoma 2 |
| chr6_+_129385816 | 0.10 |
ENSMUST00000058352.15
ENSMUST00000088075.8 |
Clec9a
|
C-type lectin domain family 9, member a |
| chr13_-_74918701 | 0.10 |
ENSMUST00000223126.2
|
Cast
|
calpastatin |
| chr13_+_99481283 | 0.10 |
ENSMUST00000052249.7
ENSMUST00000224660.3 |
Mrps27
|
mitochondrial ribosomal protein S27 |
| chr16_+_87251852 | 0.10 |
ENSMUST00000119504.8
ENSMUST00000131356.8 |
Usp16
|
ubiquitin specific peptidase 16 |
| chr11_+_96355413 | 0.09 |
ENSMUST00000103154.11
ENSMUST00000100521.10 ENSMUST00000100519.11 |
Skap1
|
src family associated phosphoprotein 1 |
| chr11_-_102120953 | 0.09 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr9_+_38630317 | 0.09 |
ENSMUST00000129598.2
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr12_+_21366386 | 0.09 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr8_-_104975134 | 0.09 |
ENSMUST00000212275.2
ENSMUST00000050211.7 |
Tk2
|
thymidine kinase 2, mitochondrial |
| chr18_-_13013030 | 0.09 |
ENSMUST00000119512.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr7_+_44033520 | 0.08 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr16_-_16687119 | 0.08 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr13_-_64514830 | 0.08 |
ENSMUST00000222971.2
|
Ctsl
|
cathepsin L |
| chr1_+_165130192 | 0.08 |
ENSMUST00000111450.3
|
Gpr161
|
G protein-coupled receptor 161 |
| chr10_-_83369994 | 0.08 |
ENSMUST00000020497.14
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr16_+_38383154 | 0.08 |
ENSMUST00000171687.8
ENSMUST00000002924.15 |
Tmem39a
|
transmembrane protein 39a |
| chr7_+_81220987 | 0.08 |
ENSMUST00000165460.2
|
Whamm
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr13_-_69147639 | 0.08 |
ENSMUST00000022013.8
|
Adcy2
|
adenylate cyclase 2 |
| chr4_-_42528175 | 0.08 |
ENSMUST00000180201.2
|
Fam205a2
|
family with sequence similarity 205, member A2 |
| chr4_-_117013396 | 0.08 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 0.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 0.4 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.5 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.4 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.3 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 1.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.7 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 0.9 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:1903232 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.3 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.2 | GO:0061034 | succinyl-CoA metabolic process(GO:0006104) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.4 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) mesonephric duct formation(GO:0072181) |
| 0.0 | 0.1 | GO:1904109 | polar body extrusion after meiotic divisions(GO:0040038) spindle assembly involved in meiosis(GO:0090306) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:1902996 | regulation of neuronal signal transduction(GO:1902847) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.4 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.2 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.3 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 0.4 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.1 | 0.7 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.2 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.2 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0051861 | glycolipid binding(GO:0051861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |