Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
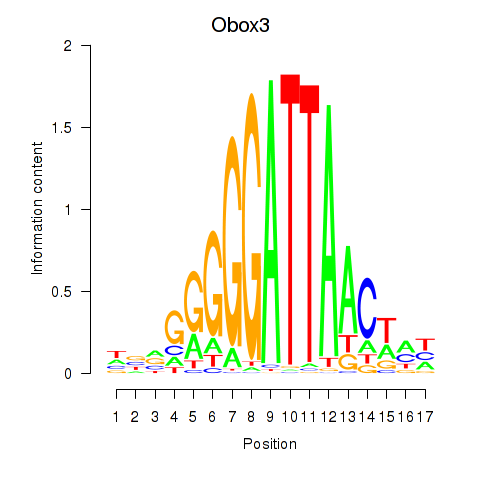
Results for Obox3
Z-value: 0.76
Transcription factors associated with Obox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Obox3
|
ENSMUSG00000066772.13 | oocyte specific homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox3 | mm39_v1_chr7_-_15361801_15361801 | -0.76 | 1.4e-01 | Click! |
Activity profile of Obox3 motif
Sorted Z-values of Obox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_109540201 | 0.58 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr11_-_68864666 | 0.51 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr4_-_15945359 | 0.39 |
ENSMUST00000029877.9
|
Decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr10_-_75617245 | 0.37 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr2_-_23939401 | 0.34 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr11_+_95275458 | 0.33 |
ENSMUST00000021243.16
ENSMUST00000146556.2 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr12_-_101943134 | 0.32 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr4_+_132495636 | 0.31 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr11_-_53321242 | 0.30 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr14_-_31362835 | 0.30 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr15_+_81548090 | 0.28 |
ENSMUST00000023029.15
ENSMUST00000174229.8 ENSMUST00000172748.8 |
L3mbtl2
|
L3MBTL2 polycomb repressive complex 1 subunit |
| chr9_+_73020708 | 0.28 |
ENSMUST00000169399.8
ENSMUST00000034738.14 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chr13_+_41154478 | 0.27 |
ENSMUST00000046951.10
|
Pak1ip1
|
PAK1 interacting protein 1 |
| chr17_+_47908025 | 0.26 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr2_-_126342551 | 0.26 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr8_+_71940747 | 0.25 |
ENSMUST00000007754.13
ENSMUST00000168847.8 |
Gtpbp3
|
GTP binding protein 3 |
| chr7_-_25488060 | 0.24 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chrX_+_73468140 | 0.23 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr9_-_21223551 | 0.23 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr7_-_25098122 | 0.23 |
ENSMUST00000105177.3
ENSMUST00000149349.2 |
Lipe
|
lipase, hormone sensitive |
| chr2_+_144435974 | 0.23 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr1_-_52271455 | 0.22 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr14_+_59438658 | 0.22 |
ENSMUST00000173547.8
ENSMUST00000043227.13 ENSMUST00000022551.14 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr5_-_65248927 | 0.21 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr14_-_20027112 | 0.20 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_+_48438751 | 0.19 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr7_-_44635740 | 0.17 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr9_+_38630317 | 0.17 |
ENSMUST00000129598.2
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr10_-_7556881 | 0.15 |
ENSMUST00000159977.2
ENSMUST00000162682.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr16_+_33614378 | 0.15 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr3_-_63872189 | 0.13 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr5_-_65248962 | 0.13 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr14_-_70043079 | 0.11 |
ENSMUST00000022665.4
|
Rhobtb2
|
Rho-related BTB domain containing 2 |
| chr11_+_48977495 | 0.11 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr15_+_76579960 | 0.10 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr7_-_44635813 | 0.10 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr11_+_69886652 | 0.10 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr9_+_62249730 | 0.09 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chrX_+_141010919 | 0.09 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr3_-_63872079 | 0.08 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr14_+_61375893 | 0.07 |
ENSMUST00000089394.10
|
Sacs
|
sacsin |
| chr9_-_21223631 | 0.06 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr1_+_60948149 | 0.06 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr9_+_50528608 | 0.06 |
ENSMUST00000000171.15
ENSMUST00000151197.8 |
Pih1d2
|
PIH1 domain containing 2 |
| chr3_+_55023594 | 0.05 |
ENSMUST00000146109.2
|
Spg20
|
spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
| chr16_+_32066037 | 0.05 |
ENSMUST00000141820.8
ENSMUST00000178573.8 ENSMUST00000023474.4 ENSMUST00000135289.2 |
Wdr53
|
WD repeat domain 53 |
| chr2_+_152804405 | 0.05 |
ENSMUST00000099197.9
ENSMUST00000103155.10 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr10_-_129023288 | 0.03 |
ENSMUST00000072976.4
|
Olfr773
|
olfactory receptor 773 |
| chr1_+_178015287 | 0.02 |
ENSMUST00000159284.2
|
Desi2
|
desumoylating isopeptidase 2 |
| chr9_+_39578501 | 0.02 |
ENSMUST00000215649.2
|
Olfr963
|
olfactory receptor 963 |
| chr17_+_9207180 | 0.02 |
ENSMUST00000151609.2
ENSMUST00000232775.2 ENSMUST00000136954.3 |
1700010I14Rik
|
RIKEN cDNA 1700010I14 gene |
| chr1_+_60948307 | 0.02 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr10_+_129866217 | 0.02 |
ENSMUST00000205181.3
ENSMUST00000214177.2 |
Olfr821
|
olfactory receptor 821 |
| chr7_+_88079534 | 0.02 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chr2_+_136734088 | 0.01 |
ENSMUST00000099311.9
|
Slx4ip
|
SLX4 interacting protein |
| chr4_+_80752360 | 0.01 |
ENSMUST00000133655.8
ENSMUST00000006151.13 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr2_+_67276338 | 0.01 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr4_+_80752535 | 0.01 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr14_-_20027219 | 0.01 |
ENSMUST00000055100.14
ENSMUST00000162425.8 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_-_28819331 | 0.01 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr11_+_69886603 | 0.01 |
ENSMUST00000018716.10
|
Phf23
|
PHD finger protein 23 |
| chr12_+_78243846 | 0.01 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr18_-_43526411 | 0.01 |
ENSMUST00000025379.14
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr14_-_118289557 | 0.01 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr12_+_21366386 | 0.01 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr9_+_62249341 | 0.01 |
ENSMUST00000135395.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr6_+_90246088 | 0.01 |
ENSMUST00000058039.3
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr6_-_69553484 | 0.01 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr18_-_3281752 | 0.01 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr7_+_114318746 | 0.00 |
ENSMUST00000182044.2
|
Calcb
|
calcitonin-related polypeptide, beta |
| chr7_+_107966697 | 0.00 |
ENSMUST00000210291.2
|
Olfr494
|
olfactory receptor 494 |
| chr1_+_140173787 | 0.00 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr6_+_30541581 | 0.00 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.5 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.2 | GO:0097350 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |