Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
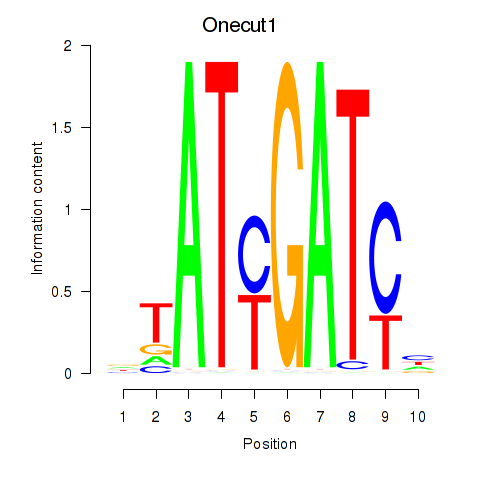
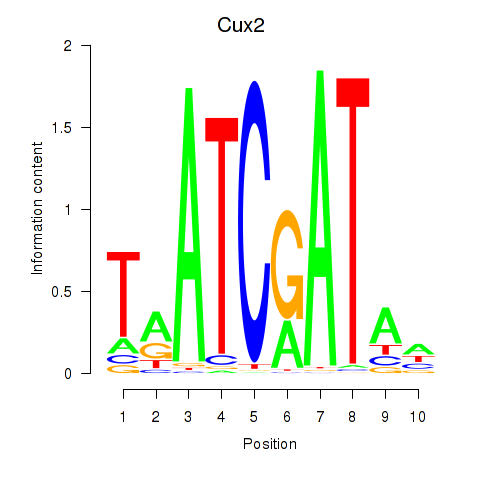
Results for Onecut1_Cux2
Z-value: 0.44
Transcription factors associated with Onecut1_Cux2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut1
|
ENSMUSG00000043013.11 | one cut domain, family member 1 |
|
Cux2
|
ENSMUSG00000042589.19 | cut-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cux2 | mm39_v1_chr5_-_122187884_122187947 | 0.90 | 3.7e-02 | Click! |
| Onecut1 | mm39_v1_chr9_+_74769166_74769203 | 0.69 | 2.0e-01 | Click! |
Activity profile of Onecut1_Cux2 motif
Sorted Z-values of Onecut1_Cux2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_172525613 | 0.53 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr15_-_76093256 | 0.39 |
ENSMUST00000071869.12
ENSMUST00000170915.2 |
Plec
|
plectin |
| chr9_-_29874401 | 0.21 |
ENSMUST00000075069.11
|
Ntm
|
neurotrimin |
| chr13_-_54836059 | 0.20 |
ENSMUST00000122935.2
ENSMUST00000128257.8 |
Rnf44
|
ring finger protein 44 |
| chrM_+_14138 | 0.20 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_-_14061 | 0.20 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr9_-_50650663 | 0.19 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr10_-_128505096 | 0.18 |
ENSMUST00000238610.2
ENSMUST00000238712.2 |
Ikzf4
|
IKAROS family zinc finger 4 |
| chr1_-_158785937 | 0.18 |
ENSMUST00000159861.9
|
Pappa2
|
pappalysin 2 |
| chr8_-_13250535 | 0.18 |
ENSMUST00000165605.4
ENSMUST00000209691.2 ENSMUST00000211128.2 ENSMUST00000210317.2 |
Grtp1
|
GH regulated TBC protein 1 |
| chr3_+_28752050 | 0.17 |
ENSMUST00000029240.14
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chrX_-_48980360 | 0.17 |
ENSMUST00000217355.3
|
Olfr1322
|
olfactory receptor 1322 |
| chr8_+_56747613 | 0.17 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr8_-_32408380 | 0.17 |
ENSMUST00000208497.3
ENSMUST00000207584.3 |
Nrg1
|
neuregulin 1 |
| chr3_+_101917455 | 0.16 |
ENSMUST00000066187.6
ENSMUST00000198675.2 |
Nhlh2
|
nescient helix loop helix 2 |
| chr15_+_54975713 | 0.15 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr11_+_97206542 | 0.15 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr4_-_104733580 | 0.14 |
ENSMUST00000064873.9
ENSMUST00000106808.10 ENSMUST00000048947.15 |
C8a
|
complement component 8, alpha polypeptide |
| chr2_-_60503998 | 0.11 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr3_-_79439181 | 0.11 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr12_-_46865709 | 0.11 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr2_-_7086018 | 0.10 |
ENSMUST00000114923.3
ENSMUST00000182706.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr15_+_54975814 | 0.10 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr18_-_84607615 | 0.10 |
ENSMUST00000125763.3
|
Zfp407
|
zinc finger protein 407 |
| chr2_+_90575714 | 0.10 |
ENSMUST00000238890.2
ENSMUST00000013759.6 |
Fnbp4
|
formin binding protein 4 |
| chr11_+_29323618 | 0.09 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr5_-_136596094 | 0.09 |
ENSMUST00000175975.9
ENSMUST00000176216.9 ENSMUST00000176745.8 |
Cux1
|
cut-like homeobox 1 |
| chr11_+_29324348 | 0.09 |
ENSMUST00000239407.2
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr12_-_25146078 | 0.09 |
ENSMUST00000222667.2
ENSMUST00000020974.7 |
Id2
|
inhibitor of DNA binding 2 |
| chrX_+_40490005 | 0.08 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr7_-_119461027 | 0.07 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr5_-_5744559 | 0.07 |
ENSMUST00000115426.9
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr10_-_63926044 | 0.07 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr6_+_121323577 | 0.06 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr14_-_55329734 | 0.06 |
ENSMUST00000036328.9
|
Zfhx2
|
zinc finger homeobox 2 |
| chrX_-_140979913 | 0.05 |
ENSMUST00000042530.4
|
Gucy2f
|
guanylate cyclase 2f |
| chr18_+_36098090 | 0.05 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr2_-_84605732 | 0.05 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr10_-_128237087 | 0.05 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr2_-_34262012 | 0.05 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr11_+_110914678 | 0.05 |
ENSMUST00000150902.8
ENSMUST00000178798.2 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr4_+_109200225 | 0.04 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr5_-_3646540 | 0.04 |
ENSMUST00000042753.14
|
Rbm48
|
RNA binding motif protein 48 |
| chr5_-_122187884 | 0.04 |
ENSMUST00000111752.10
|
Cux2
|
cut-like homeobox 2 |
| chr8_+_127790772 | 0.04 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr12_+_55646209 | 0.04 |
ENSMUST00000051857.5
|
Insm2
|
insulinoma-associated 2 |
| chr2_+_163444248 | 0.04 |
ENSMUST00000152135.8
|
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr1_-_131161312 | 0.04 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chrX_+_133305291 | 0.03 |
ENSMUST00000113228.8
ENSMUST00000153424.8 |
Drp2
|
dystrophin related protein 2 |
| chr2_+_147206910 | 0.03 |
ENSMUST00000109968.3
|
Pax1
|
paired box 1 |
| chr5_-_122188165 | 0.03 |
ENSMUST00000154139.8
|
Cux2
|
cut-like homeobox 2 |
| chr6_-_136899167 | 0.02 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr2_-_84605764 | 0.02 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr11_+_78389913 | 0.02 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr9_-_114673158 | 0.02 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr18_+_69652751 | 0.02 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chr7_+_67297152 | 0.02 |
ENSMUST00000032774.16
ENSMUST00000107471.8 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_-_7086066 | 0.02 |
ENSMUST00000183209.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr14_+_69266566 | 0.02 |
ENSMUST00000014957.10
|
Stc1
|
stanniocalcin 1 |
| chr5_+_144037171 | 0.02 |
ENSMUST00000041804.8
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr2_-_92222979 | 0.01 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr3_+_60436570 | 0.01 |
ENSMUST00000192607.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr19_-_37155410 | 0.01 |
ENSMUST00000133988.2
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_-_114023935 | 0.01 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr6_-_23132977 | 0.01 |
ENSMUST00000031707.14
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr10_-_117074501 | 0.01 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr11_-_24025054 | 0.01 |
ENSMUST00000068360.2
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr8_+_23114035 | 0.01 |
ENSMUST00000033936.8
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr19_+_31060237 | 0.01 |
ENSMUST00000066039.8
|
Cstf2t
|
cleavage stimulation factor, 3' pre-RNA subunit 2, tau |
| chr14_-_30665232 | 0.01 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr8_+_114369838 | 0.01 |
ENSMUST00000095173.3
ENSMUST00000034219.12 ENSMUST00000212269.2 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr19_+_4042752 | 0.01 |
ENSMUST00000041871.9
|
Tbx10
|
T-box 10 |
| chr4_-_46991842 | 0.01 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr7_+_45276906 | 0.01 |
ENSMUST00000057927.10
|
Rasip1
|
Ras interacting protein 1 |
| chr5_+_92831150 | 0.01 |
ENSMUST00000113055.9
|
Shroom3
|
shroom family member 3 |
| chr3_+_125197722 | 0.01 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr1_-_13197387 | 0.01 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14 |
| chr5_+_63806451 | 0.01 |
ENSMUST00000159584.3
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr9_+_59496571 | 0.00 |
ENSMUST00000121266.8
ENSMUST00000118164.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr1_+_88334678 | 0.00 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr13_+_27278173 | 0.00 |
ENSMUST00000225330.2
ENSMUST00000082079.5 |
Prl3d1
|
prolactin family 3, subfamily d, member 1 |
| chr19_+_55882942 | 0.00 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chrX_-_50294867 | 0.00 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr4_+_117706390 | 0.00 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr18_+_57601541 | 0.00 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr7_-_140859034 | 0.00 |
ENSMUST00000211667.2
ENSMUST00000167790.3 ENSMUST00000046156.13 |
Sct
|
secretin |
| chr18_+_37063237 | 0.00 |
ENSMUST00000193839.6
ENSMUST00000070797.7 |
Pcdha1
|
protocadherin alpha 1 |
| chr7_+_67305162 | 0.00 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_-_35287069 | 0.00 |
ENSMUST00000113009.2
ENSMUST00000113010.9 |
4930402F06Rik
|
RIKEN cDNA 4930402F06 gene |
| chr19_+_59207646 | 0.00 |
ENSMUST00000065204.8
|
Kcnk18
|
potassium channel, subfamily K, member 18 |
| chr12_+_85733415 | 0.00 |
ENSMUST00000040536.6
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr5_-_5744326 | 0.00 |
ENSMUST00000148333.8
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr7_+_104705455 | 0.00 |
ENSMUST00000098161.2
|
Olfr677
|
olfactory receptor 677 |
| chr19_-_12051997 | 0.00 |
ENSMUST00000087828.2
|
Olfr1425
|
olfactory receptor 1425 |
| chr11_-_55931811 | 0.00 |
ENSMUST00000037682.3
|
Nmur2
|
neuromedin U receptor 2 |
| chrX_+_100420873 | 0.00 |
ENSMUST00000052130.14
|
Gjb1
|
gap junction protein, beta 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Onecut1_Cux2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.2 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |