Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
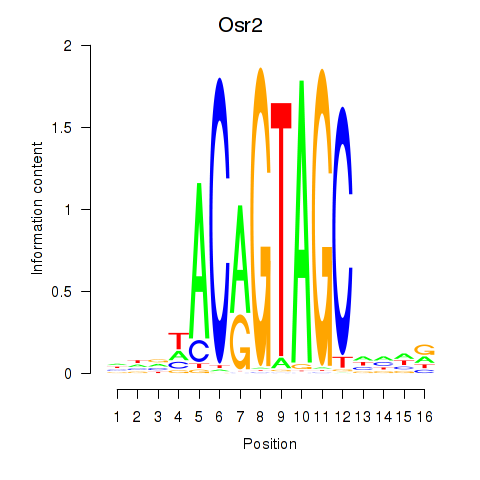
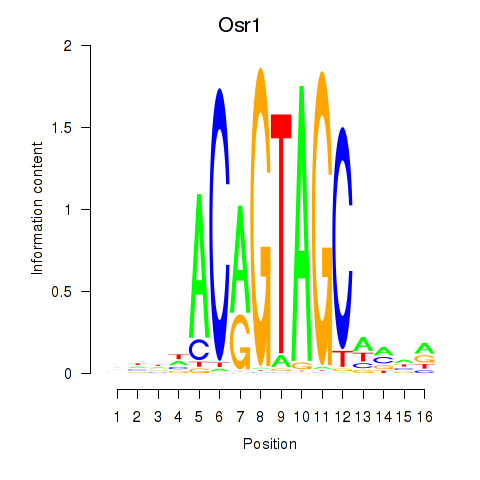
Results for Osr2_Osr1
Z-value: 0.03
Transcription factors associated with Osr2_Osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Osr2
|
ENSMUSG00000022330.6 | odd-skipped related 2 |
|
Osr1
|
ENSMUSG00000048387.9 | odd-skipped related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Osr2 | mm39_v1_chr15_+_35296237_35296250 | 0.63 | 2.6e-01 | Click! |
| Osr1 | mm39_v1_chr12_+_9624437_9624448 | -0.09 | 8.9e-01 | Click! |
Activity profile of Osr2_Osr1 motif
Sorted Z-values of Osr2_Osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_3671528 | 0.07 |
ENSMUST00000156380.4
|
Pet100
|
PET100 homolog |
| chr7_-_127490139 | 0.07 |
ENSMUST00000205300.2
ENSMUST00000121394.3 |
Prss53
|
protease, serine 53 |
| chr5_-_113957362 | 0.07 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr1_-_192880260 | 0.06 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr4_+_129355374 | 0.05 |
ENSMUST00000048162.10
ENSMUST00000138013.3 |
Bsdc1
|
BSD domain containing 1 |
| chr19_+_5012336 | 0.05 |
ENSMUST00000237974.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr14_-_20529997 | 0.05 |
ENSMUST00000225132.2
|
Anxa7
|
annexin A7 |
| chr19_+_7534816 | 0.04 |
ENSMUST00000136465.8
ENSMUST00000025925.11 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr2_-_69715899 | 0.04 |
ENSMUST00000060447.13
|
Mettl5
|
methyltransferase like 5 |
| chr13_-_100589357 | 0.04 |
ENSMUST00000222155.2
ENSMUST00000221727.2 |
Naip1
|
NLR family, apoptosis inhibitory protein 1 |
| chr7_-_123099672 | 0.04 |
ENSMUST00000042470.14
ENSMUST00000128217.2 |
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr4_+_130640611 | 0.04 |
ENSMUST00000156225.8
ENSMUST00000156742.8 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr2_+_179713586 | 0.04 |
ENSMUST00000108901.8
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr6_+_122929627 | 0.04 |
ENSMUST00000204427.2
|
Clec4a3
|
C-type lectin domain family 4, member a3 |
| chr11_+_69882410 | 0.03 |
ENSMUST00000108592.2
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr13_+_23935088 | 0.03 |
ENSMUST00000078369.3
|
H2ac4
|
H2A clustered histone 4 |
| chr9_-_58648826 | 0.03 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr17_+_57182472 | 0.03 |
ENSMUST00000025048.7
|
Acsbg3
|
acyl-CoA synthetase bubblegum family member 3 |
| chr7_-_6511472 | 0.03 |
ENSMUST00000207055.3
ENSMUST00000209097.2 ENSMUST00000208623.3 ENSMUST00000208207.2 ENSMUST00000209029.4 ENSMUST00000214383.2 ENSMUST00000207624.2 ENSMUST00000213549.2 |
Olfr1348
|
olfactory receptor 1348 |
| chr15_-_103446354 | 0.03 |
ENSMUST00000023133.8
|
Ppp1r1a
|
protein phosphatase 1, regulatory inhibitor subunit 1A |
| chr2_-_86180622 | 0.02 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr10_+_3690348 | 0.02 |
ENSMUST00000120274.8
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr3_-_127019496 | 0.02 |
ENSMUST00000182064.9
ENSMUST00000182452.8 |
Ank2
|
ankyrin 2, brain |
| chr12_-_108668520 | 0.02 |
ENSMUST00000167978.9
ENSMUST00000021691.6 |
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr8_-_23143422 | 0.02 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr18_-_36899245 | 0.02 |
ENSMUST00000061522.8
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr1_+_88066086 | 0.02 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr6_-_119940694 | 0.02 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr7_-_30443106 | 0.02 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr2_-_36372480 | 0.02 |
ENSMUST00000213300.2
|
Olfr341
|
olfactory receptor 341 |
| chr11_+_72192455 | 0.02 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr1_-_23436620 | 0.02 |
ENSMUST00000188677.2
|
Ogfrl1
|
opioid growth factor receptor-like 1 |
| chr5_+_145217272 | 0.02 |
ENSMUST00000200246.2
|
Zscan25
|
zinc finger and SCAN domain containing 25 |
| chr3_+_87343071 | 0.02 |
ENSMUST00000194102.6
|
Fcrl5
|
Fc receptor-like 5 |
| chr4_-_116413092 | 0.01 |
ENSMUST00000069674.6
ENSMUST00000106478.9 |
Tmem69
|
transmembrane protein 69 |
| chr14_+_77274185 | 0.01 |
ENSMUST00000048208.10
ENSMUST00000095625.11 |
Ccdc122
|
coiled-coil domain containing 122 |
| chr13_+_74498430 | 0.01 |
ENSMUST00000160021.8
ENSMUST00000162672.8 ENSMUST00000162376.2 |
Ccdc127
|
coiled-coil domain containing 127 |
| chr5_+_107655487 | 0.01 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr14_-_51493569 | 0.01 |
ENSMUST00000061936.8
|
Rnase2a
|
ribonuclease, RNase A family, 2A (liver, eosinophil-derived neurotoxin) |
| chr19_+_37423198 | 0.01 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr2_+_172090412 | 0.01 |
ENSMUST00000038532.2
|
Mc3r
|
melanocortin 3 receptor |
| chr16_+_91188609 | 0.01 |
ENSMUST00000160764.2
|
Gm21970
|
predicted gene 21970 |
| chr6_+_42716305 | 0.01 |
ENSMUST00000213997.2
|
Olfr453
|
olfactory receptor 453 |
| chr16_-_58712421 | 0.01 |
ENSMUST00000206523.3
ENSMUST00000215032.2 |
Olfr178
|
olfactory receptor 178 |
| chr8_-_46428277 | 0.01 |
ENSMUST00000095323.8
ENSMUST00000098786.3 |
1700029J07Rik
|
RIKEN cDNA 1700029J07 gene |
| chr14_+_50360643 | 0.01 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr2_+_37029334 | 0.01 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chrX_-_133062677 | 0.01 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr9_+_45818250 | 0.01 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr1_-_74640504 | 0.01 |
ENSMUST00000136078.2
ENSMUST00000132081.2 ENSMUST00000113721.8 ENSMUST00000027357.12 |
Rnf25
|
ring finger protein 25 |
| chr1_-_156248716 | 0.01 |
ENSMUST00000178036.8
|
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr7_+_19699291 | 0.01 |
ENSMUST00000094753.6
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chrX_+_36007312 | 0.01 |
ENSMUST00000047655.7
|
Slc25a43
|
solute carrier family 25, member 43 |
| chr15_+_84116231 | 0.01 |
ENSMUST00000023072.7
|
Parvb
|
parvin, beta |
| chr2_-_177567397 | 0.01 |
ENSMUST00000108934.9
ENSMUST00000081529.11 |
Zfp972
|
zinc finger protein 972 |
| chr15_+_35371644 | 0.01 |
ENSMUST00000227455.2
|
Vps13b
|
vacuolar protein sorting 13B |
| chr9_-_35179042 | 0.01 |
ENSMUST00000217306.2
ENSMUST00000125087.2 ENSMUST00000121564.8 ENSMUST00000063782.12 ENSMUST00000059057.14 |
Fam118b
|
family with sequence similarity 118, member B |
| chr19_+_18609343 | 0.01 |
ENSMUST00000159572.9
|
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr11_+_119804630 | 0.01 |
ENSMUST00000026434.13
ENSMUST00000124199.8 |
Chmp6
|
charged multivesicular body protein 6 |
| chrX_-_7185529 | 0.01 |
ENSMUST00000128319.2
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr9_+_45817795 | 0.01 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr17_+_38112070 | 0.01 |
ENSMUST00000217365.2
|
Olfr124
|
olfactory receptor 124 |
| chr12_-_110649040 | 0.01 |
ENSMUST00000222915.2
ENSMUST00000070659.7 |
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr9_-_55190902 | 0.01 |
ENSMUST00000164721.8
|
Nrg4
|
neuregulin 4 |
| chr7_-_24459736 | 0.01 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr2_-_5719302 | 0.01 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr11_-_102446947 | 0.01 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8 |
| chr1_-_85888729 | 0.01 |
ENSMUST00000086975.7
|
Gpr55
|
G protein-coupled receptor 55 |
| chr5_-_110596387 | 0.01 |
ENSMUST00000198768.3
|
Fbrsl1
|
fibrosin-like 1 |
| chr19_+_5927876 | 0.01 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr9_-_96601574 | 0.01 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr10_+_120062954 | 0.01 |
ENSMUST00000020444.16
|
Llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr13_-_4329421 | 0.01 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr13_+_74498418 | 0.01 |
ENSMUST00000022063.14
|
Ccdc127
|
coiled-coil domain containing 127 |
| chr14_-_31157985 | 0.01 |
ENSMUST00000091903.5
|
Sh3bp5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr15_+_35371300 | 0.01 |
ENSMUST00000048646.9
|
Vps13b
|
vacuolar protein sorting 13B |
| chrX_-_166047289 | 0.01 |
ENSMUST00000133722.2
|
Tlr8
|
toll-like receptor 8 |
| chr11_+_51111300 | 0.01 |
ENSMUST00000126189.3
|
Msantd5
|
Myb/SANT DNA binding domain containing 5 |
| chrY_-_6681243 | 0.01 |
ENSMUST00000115940.2
|
Gm21719
|
predicted gene, 21719 |
| chr11_-_70130620 | 0.01 |
ENSMUST00000040428.4
|
Rnasek
|
ribonuclease, RNase K |
| chr12_-_55061117 | 0.01 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chrX_-_7185424 | 0.01 |
ENSMUST00000115746.8
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr12_+_112645237 | 0.01 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr5_-_66238313 | 0.01 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr9_-_39465349 | 0.01 |
ENSMUST00000215505.2
ENSMUST00000217227.2 |
Olfr958
|
olfactory receptor 958 |
| chr6_+_37877413 | 0.01 |
ENSMUST00000120238.2
|
Trim24
|
tripartite motif-containing 24 |
| chr2_+_176522933 | 0.01 |
ENSMUST00000126726.2
ENSMUST00000122218.9 |
Gm14408
|
predicted gene 14408 |
| chr4_+_45890303 | 0.01 |
ENSMUST00000178561.8
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr14_+_120513106 | 0.01 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr8_+_27513839 | 0.01 |
ENSMUST00000209563.2
ENSMUST00000209520.2 |
Erlin2
|
ER lipid raft associated 2 |
| chrX_-_166047275 | 0.01 |
ENSMUST00000112170.2
|
Tlr8
|
toll-like receptor 8 |
| chr7_+_126895531 | 0.01 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr19_-_14575395 | 0.01 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr7_+_45175754 | 0.01 |
ENSMUST00000211227.2
ENSMUST00000051810.15 |
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr13_+_20978283 | 0.01 |
ENSMUST00000021757.5
ENSMUST00000221982.2 |
Aoah
|
acyloxyacyl hydrolase |
| chr3_+_90520176 | 0.01 |
ENSMUST00000001051.9
|
S100a6
|
S100 calcium binding protein A6 (calcyclin) |
| chr19_-_18609118 | 0.00 |
ENSMUST00000025631.7
ENSMUST00000236615.2 |
Ostf1
|
osteoclast stimulating factor 1 |
| chr2_+_136734088 | 0.00 |
ENSMUST00000099311.9
|
Slx4ip
|
SLX4 interacting protein |
| chr12_-_113700190 | 0.00 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr1_+_10108433 | 0.00 |
ENSMUST00000071087.12
ENSMUST00000117415.8 |
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr4_+_125997788 | 0.00 |
ENSMUST00000139524.2
|
Stk40
|
serine/threonine kinase 40 |
| chr7_+_141503719 | 0.00 |
ENSMUST00000105989.9
ENSMUST00000075528.12 ENSMUST00000174499.8 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr7_+_96600756 | 0.00 |
ENSMUST00000107159.3
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr9_-_110483210 | 0.00 |
ENSMUST00000196488.5
ENSMUST00000133191.8 ENSMUST00000167320.8 |
Nbeal2
|
neurobeachin-like 2 |
| chr9_-_50650663 | 0.00 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr9_-_110571645 | 0.00 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr3_-_95264467 | 0.00 |
ENSMUST00000107171.10
ENSMUST00000015841.12 ENSMUST00000107170.3 |
Setdb1
|
SET domain, bifurcated 1 |
| chr9_-_7184440 | 0.00 |
ENSMUST00000140466.8
|
Dync2h1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr6_+_67586695 | 0.00 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr15_-_89314054 | 0.00 |
ENSMUST00000023289.13
|
Chkb
|
choline kinase beta |
| chr1_+_171097891 | 0.00 |
ENSMUST00000064272.10
ENSMUST00000151863.8 ENSMUST00000141999.8 ENSMUST00000111313.10 ENSMUST00000126699.4 |
B4galt3
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 3 |
| chr3_+_145281941 | 0.00 |
ENSMUST00000199033.5
ENSMUST00000098534.9 ENSMUST00000200574.5 ENSMUST00000196413.5 ENSMUST00000197604.3 |
Znhit6
|
zinc finger, HIT type 6 |
| chr1_-_80642969 | 0.00 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr5_-_148931957 | 0.00 |
ENSMUST00000147473.6
|
Gm42791
|
predicted gene 42791 |
| chr9_-_42372710 | 0.00 |
ENSMUST00000066179.14
|
Tbcel
|
tubulin folding cofactor E-like |
| chr7_+_44499005 | 0.00 |
ENSMUST00000150335.2
ENSMUST00000107882.8 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr5_+_124068728 | 0.00 |
ENSMUST00000094320.10
ENSMUST00000165148.4 |
Ccdc62
|
coiled-coil domain containing 62 |
| chr11_-_98040377 | 0.00 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr19_+_18609291 | 0.00 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr9_-_45818134 | 0.00 |
ENSMUST00000161203.8
ENSMUST00000058720.13 |
Rnf214
|
ring finger protein 214 |
| chr13_-_63545981 | 0.00 |
ENSMUST00000160735.8
|
Fancc
|
Fanconi anemia, complementation group C |
| chrX_+_55500170 | 0.00 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr16_-_30129591 | 0.00 |
ENSMUST00000061190.8
|
Gp5
|
glycoprotein 5 (platelet) |
| chr7_+_24920840 | 0.00 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr14_+_44340111 | 0.00 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr10_+_43777777 | 0.00 |
ENSMUST00000054418.12
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr19_-_47303184 | 0.00 |
ENSMUST00000237796.2
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr6_+_117883732 | 0.00 |
ENSMUST00000179224.8
ENSMUST00000035493.14 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chrX_-_101730578 | 0.00 |
ENSMUST00000122154.3
|
Gm3880
|
predicted pseudogene 3880 |
| chr1_+_90131702 | 0.00 |
ENSMUST00000065587.5
ENSMUST00000159654.2 |
Ackr3
|
atypical chemokine receptor 3 |
| chr16_+_45430743 | 0.00 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr8_+_27513819 | 0.00 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr10_-_57408585 | 0.00 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr12_+_52144511 | 0.00 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr3_+_87343105 | 0.00 |
ENSMUST00000193229.6
|
Fcrl5
|
Fc receptor-like 5 |
| chr17_+_8454862 | 0.00 |
ENSMUST00000231340.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr5_-_145406533 | 0.00 |
ENSMUST00000031633.5
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr13_+_92442488 | 0.00 |
ENSMUST00000188317.2
|
Gm20379
|
predicted gene, 20379 |
| chr6_+_90346468 | 0.00 |
ENSMUST00000113539.8
ENSMUST00000075117.10 |
Zxdc
|
ZXD family zinc finger C |
| chr8_+_22108199 | 0.00 |
ENSMUST00000074343.6
|
Defa26
|
defensin, alpha, 26 |
| chr9_-_54100726 | 0.00 |
ENSMUST00000034811.8
ENSMUST00000215736.2 |
Cyp19a1
|
cytochrome P450, family 19, subfamily a, polypeptide 1 |
| chr9_+_75255037 | 0.00 |
ENSMUST00000034709.7
|
Bcl2l10
|
Bcl2-like 10 |
| chr5_-_49442980 | 0.00 |
ENSMUST00000175660.5
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr16_-_20972750 | 0.00 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr9_-_45818196 | 0.00 |
ENSMUST00000160699.9
|
Rnf214
|
ring finger protein 214 |
| chr19_-_29721012 | 0.00 |
ENSMUST00000175764.9
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr9_+_51124983 | 0.00 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr7_+_141503411 | 0.00 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr6_-_69741999 | 0.00 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr4_-_138641225 | 0.00 |
ENSMUST00000097830.4
|
Otud3
|
OTU domain containing 3 |
| chr5_+_9316097 | 0.00 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr16_-_10608766 | 0.00 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr10_+_84674008 | 0.00 |
ENSMUST00000095388.5
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr14_-_20529974 | 0.00 |
ENSMUST00000224975.2
|
Anxa7
|
annexin A7 |
| chr19_-_16851169 | 0.00 |
ENSMUST00000072915.4
|
Foxb2
|
forkhead box B2 |
| chrX_+_106711562 | 0.00 |
ENSMUST00000168174.9
|
Tbx22
|
T-box 22 |
| chr19_-_12868022 | 0.00 |
ENSMUST00000081236.3
|
Olfr1446
|
olfactory receptor 1446 |
| chr8_-_68574179 | 0.00 |
ENSMUST00000178529.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_+_48096628 | 0.00 |
ENSMUST00000024786.14
|
Tfeb
|
transcription factor EB |
| chr18_+_34675366 | 0.00 |
ENSMUST00000012426.3
|
Wnt8a
|
wingless-type MMTV integration site family, member 8A |
| chr15_-_36140539 | 0.00 |
ENSMUST00000172831.8
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr3_-_89230190 | 0.00 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr13_-_40887244 | 0.00 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr11_-_100650768 | 0.00 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr8_-_25085654 | 0.00 |
ENSMUST00000110667.8
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr11_-_100650566 | 0.00 |
ENSMUST00000107361.9
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chrX_-_135369790 | 0.00 |
ENSMUST00000072125.6
|
Kir3dl2
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 2 |
| chr9_-_51240201 | 0.00 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr3_+_87343127 | 0.00 |
ENSMUST00000166297.7
ENSMUST00000049926.15 ENSMUST00000178261.3 |
Fcrl5
|
Fc receptor-like 5 |
| chr7_-_104383569 | 0.00 |
ENSMUST00000211596.2
|
Olfr664
|
olfactory receptor 664 |
| chr3_+_88744323 | 0.00 |
ENSMUST00000081695.14
ENSMUST00000090942.6 |
Gon4l
|
gon-4-like (C.elegans) |
| chr15_+_76832681 | 0.00 |
ENSMUST00000071792.7
ENSMUST00000230274.2 |
1110038F14Rik
|
RIKEN cDNA 1110038F14 gene |
| chr9_-_42035560 | 0.00 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr11_+_8998575 | 0.00 |
ENSMUST00000043285.5
|
Gm11992
|
predicted gene 11992 |
| chr2_+_34764496 | 0.00 |
ENSMUST00000028228.6
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr9_-_109233364 | 0.00 |
ENSMUST00000080626.9
|
Fbxw22
|
F-box and WD-40 domain protein 22 |
| chr10_-_80374916 | 0.00 |
ENSMUST00000219648.2
|
Atp8b3
|
ATPase, class I, type 8B, member 3 |
| chr15_-_59888446 | 0.00 |
ENSMUST00000096421.4
|
4933412E24Rik
|
RIKEN cDNA 4933412E24 gene |
| chr6_-_86710220 | 0.00 |
ENSMUST00000113679.2
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr2_+_34764408 | 0.00 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr8_+_21881827 | 0.00 |
ENSMUST00000120874.5
|
Defa33
|
defensin, alpha, 33 |
| chr9_+_48406706 | 0.00 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr3_+_93107517 | 0.00 |
ENSMUST00000098884.4
|
Flg2
|
filaggrin family member 2 |
| chr4_+_125997734 | 0.00 |
ENSMUST00000116286.9
ENSMUST00000094761.11 |
Stk40
|
serine/threonine kinase 40 |
| chr10_-_109669053 | 0.00 |
ENSMUST00000238286.2
|
Nav3
|
neuron navigator 3 |
| chr7_+_10180716 | 0.00 |
ENSMUST00000055964.8
|
Vmn1r67
|
vomeronasal 1 receptor 67 |
| chr1_+_187995096 | 0.00 |
ENSMUST00000060479.14
|
Ush2a
|
usherin |
| chr9_-_16289527 | 0.00 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr6_+_141470105 | 0.00 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr7_-_57159743 | 0.00 |
ENSMUST00000068456.8
ENSMUST00000206734.2 |
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr1_+_20801127 | 0.00 |
ENSMUST00000027061.5
|
Il17a
|
interleukin 17A |
| chr6_+_83985495 | 0.00 |
ENSMUST00000113821.8
|
Dysf
|
dysferlin |
| chr4_-_58499398 | 0.00 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr1_+_74640590 | 0.00 |
ENSMUST00000087183.11
ENSMUST00000148456.8 ENSMUST00000113694.8 |
Stk36
|
serine/threonine kinase 36 |
| chr3_-_98670369 | 0.00 |
ENSMUST00000107019.8
ENSMUST00000107018.8 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chrY_+_5944438 | 0.00 |
ENSMUST00000178779.2
|
Gm21778
|
predicted gene, 21778 |
| chr8_+_21515561 | 0.00 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr4_-_133981387 | 0.00 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr2_-_19558719 | 0.00 |
ENSMUST00000062060.5
|
4921504E06Rik
|
RIKEN cDNA 4921504E06 gene |
| chr16_+_29884153 | 0.00 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Osr2_Osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |