Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
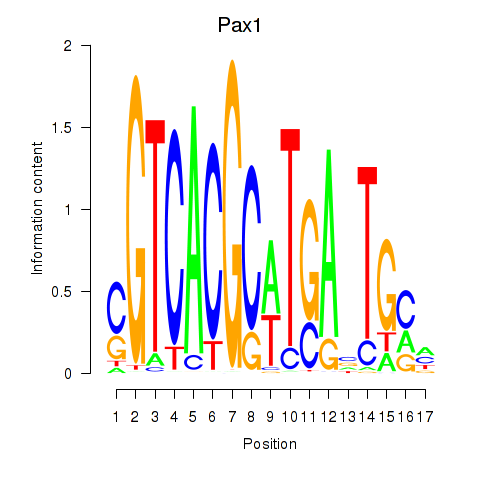
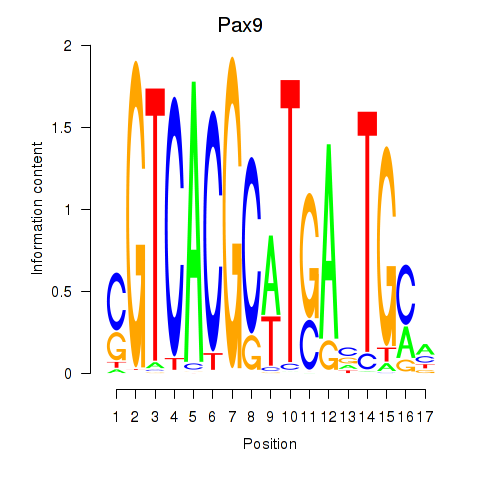
Results for Pax1_Pax9
Z-value: 0.64
Transcription factors associated with Pax1_Pax9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax1
|
ENSMUSG00000037034.17 | paired box 1 |
|
Pax9
|
ENSMUSG00000001497.19 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax1 | mm39_v1_chr2_+_147206910_147206931 | -0.67 | 2.2e-01 | Click! |
| Pax9 | mm39_v1_chr12_+_56742413_56742459 | -0.50 | 3.9e-01 | Click! |
Activity profile of Pax1_Pax9 motif
Sorted Z-values of Pax1_Pax9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax1_Pax9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |