Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Pax4
Z-value: 0.91
Transcription factors associated with Pax4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax4
|
ENSMUSG00000029706.16 | paired box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax4 | mm39_v1_chr6_-_28449250_28449352 | 0.34 | 5.7e-01 | Click! |
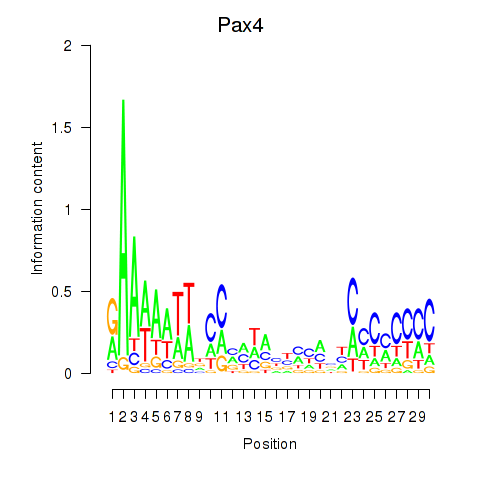
Activity profile of Pax4 motif
Sorted Z-values of Pax4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_21901791 | 1.38 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr13_+_23922783 | 1.04 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chrM_+_3906 | 0.90 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr12_+_33364288 | 0.82 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr5_-_116427003 | 0.61 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr7_+_102246977 | 0.57 |
ENSMUST00000215712.2
|
Olfr552
|
olfactory receptor 552 |
| chr9_-_48747232 | 0.56 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr13_-_40883893 | 0.51 |
ENSMUST00000021787.7
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr12_-_84265609 | 0.47 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr13_-_110416637 | 0.43 |
ENSMUST00000167824.3
ENSMUST00000224180.2 |
Rab3c
|
RAB3C, member RAS oncogene family |
| chr15_-_50753437 | 0.42 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr12_+_71095112 | 0.40 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chrM_+_11735 | 0.37 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrM_+_5319 | 0.34 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr10_+_129153986 | 0.32 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr12_-_80159768 | 0.30 |
ENSMUST00000219642.2
ENSMUST00000165114.2 ENSMUST00000218835.2 ENSMUST00000021552.3 |
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr4_+_150366028 | 0.28 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr17_+_47604995 | 0.28 |
ENSMUST00000190020.4
|
Trerf1
|
transcriptional regulating factor 1 |
| chr15_+_80861966 | 0.27 |
ENSMUST00000139517.9
ENSMUST00000137255.3 ENSMUST00000137004.2 |
Sgsm3
|
small G protein signaling modulator 3 |
| chr16_-_36486429 | 0.25 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr7_+_78563513 | 0.24 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr2_-_89491560 | 0.23 |
ENSMUST00000111527.4
|
Olfr1250
|
olfactory receptor 1250 |
| chr8_-_32499513 | 0.23 |
ENSMUST00000208205.3
|
Nrg1
|
neuregulin 1 |
| chr15_-_50753061 | 0.23 |
ENSMUST00000165201.9
ENSMUST00000184458.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chr11_-_18968955 | 0.21 |
ENSMUST00000068264.14
ENSMUST00000185131.8 |
Meis1
|
Meis homeobox 1 |
| chr1_-_155910546 | 0.21 |
ENSMUST00000169241.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr2_-_111908072 | 0.20 |
ENSMUST00000213577.2
ENSMUST00000216071.2 |
Olfr1313
|
olfactory receptor 1313 |
| chr17_+_27160203 | 0.18 |
ENSMUST00000194598.6
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chrX_-_99638466 | 0.17 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr9_+_14411907 | 0.16 |
ENSMUST00000004200.9
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr18_+_53995156 | 0.16 |
ENSMUST00000069597.8
|
Csnk1g3
|
casein kinase 1, gamma 3 |
| chr9_+_18848418 | 0.15 |
ENSMUST00000218385.2
|
Olfr832
|
olfactory receptor 832 |
| chr1_+_42734889 | 0.15 |
ENSMUST00000054883.4
|
Pou3f3
|
POU domain, class 3, transcription factor 3 |
| chr7_+_43057611 | 0.15 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr1_+_42734051 | 0.15 |
ENSMUST00000239323.2
ENSMUST00000199521.5 ENSMUST00000176807.3 |
Pou3f3
Gm20646
|
POU domain, class 3, transcription factor 3 predicted gene 20646 |
| chr10_+_127256993 | 0.15 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr11_-_18968714 | 0.15 |
ENSMUST00000177417.8
|
Meis1
|
Meis homeobox 1 |
| chr7_+_7174315 | 0.14 |
ENSMUST00000051435.8
|
Zfp418
|
zinc finger protein 418 |
| chr5_+_77099154 | 0.14 |
ENSMUST00000031160.16
ENSMUST00000120912.8 ENSMUST00000117536.8 |
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr9_+_20209828 | 0.13 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr6_+_136495818 | 0.13 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_-_83128992 | 0.13 |
ENSMUST00000210094.2
|
Il15
|
interleukin 15 |
| chr3_+_52175757 | 0.12 |
ENSMUST00000053764.7
|
Foxo1
|
forkhead box O1 |
| chr19_-_12901783 | 0.12 |
ENSMUST00000213713.2
ENSMUST00000216888.2 ENSMUST00000213177.2 |
Olfr1448
|
olfactory receptor 1448 |
| chr2_-_32976378 | 0.12 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr8_-_85500010 | 0.11 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr9_-_101076198 | 0.11 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr13_+_109769294 | 0.10 |
ENSMUST00000135275.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr11_+_106916430 | 0.10 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chr9_+_92157799 | 0.10 |
ENSMUST00000126911.2
|
Plscr2
|
phospholipid scramblase 2 |
| chr19_-_10460238 | 0.10 |
ENSMUST00000235392.2
ENSMUST00000237522.2 ENSMUST00000038842.5 |
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr9_-_58066484 | 0.10 |
ENSMUST00000041477.15
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr19_+_53128861 | 0.09 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr2_-_65068960 | 0.09 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr7_+_78563184 | 0.09 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr8_-_83129134 | 0.09 |
ENSMUST00000209363.2
|
Il15
|
interleukin 15 |
| chr3_+_95532282 | 0.08 |
ENSMUST00000058230.13
ENSMUST00000037983.6 |
Ensa
|
endosulfine alpha |
| chr2_-_65068917 | 0.08 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr7_+_24561616 | 0.07 |
ENSMUST00000170837.3
|
Gm9844
|
predicted pseudogene 9844 |
| chr17_+_34457868 | 0.06 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr8_-_22966831 | 0.06 |
ENSMUST00000163774.3
ENSMUST00000033935.16 |
Smim19
|
small integral membrane protein 19 |
| chr2_+_43445359 | 0.06 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr7_-_144292288 | 0.06 |
ENSMUST00000238848.2
ENSMUST00000118556.9 ENSMUST00000033393.15 |
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr6_+_136495784 | 0.06 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_-_59245998 | 0.05 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5 |
| chr10_+_107107477 | 0.05 |
ENSMUST00000020057.16
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr11_+_78389913 | 0.05 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr10_-_129509659 | 0.05 |
ENSMUST00000213294.2
ENSMUST00000216067.3 ENSMUST00000203424.2 |
Olfr801
|
olfactory receptor 801 |
| chr10_-_67384898 | 0.04 |
ENSMUST00000075686.7
|
Ado
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr10_-_63926044 | 0.04 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr17_+_17622934 | 0.03 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr5_+_66833434 | 0.03 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr19_-_11058452 | 0.03 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chrX_-_47966588 | 0.03 |
ENSMUST00000114928.2
|
Fsip2l
|
fibrous sheath-interacting protein 2-like |
| chr10_+_119828821 | 0.03 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr4_+_146695418 | 0.02 |
ENSMUST00000130825.8
|
Zfp993
|
zinc finger protein 993 |
| chr1_-_84912810 | 0.02 |
ENSMUST00000027422.7
|
Slc16a14
|
solute carrier family 16 (monocarboxylic acid transporters), member 14 |
| chr2_+_158509039 | 0.02 |
ENSMUST00000045503.11
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr2_+_181322077 | 0.02 |
ENSMUST00000103042.10
|
Tcea2
|
transcription elongation factor A (SII), 2 |
| chr16_-_45664591 | 0.02 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chrX_-_72140623 | 0.02 |
ENSMUST00000114524.9
ENSMUST00000074619.6 |
Xlr3a
|
X-linked lymphocyte-regulated 3A |
| chr16_-_14109219 | 0.01 |
ENSMUST00000230397.2
ENSMUST00000231567.2 ENSMUST00000090287.5 |
Myh11
|
myosin, heavy polypeptide 11, smooth muscle |
| chr7_+_23000617 | 0.01 |
ENSMUST00000076470.5
|
Nlrp4e
|
NLR family, pyrin domain containing 4E |
| chr18_+_58969739 | 0.01 |
ENSMUST00000052907.7
|
Adamts19
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 19 |
| chr7_-_144292257 | 0.01 |
ENSMUST00000121758.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr2_-_160155536 | 0.01 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr7_+_80764564 | 0.01 |
ENSMUST00000119083.2
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr17_+_27160356 | 0.01 |
ENSMUST00000229490.2
ENSMUST00000201702.5 ENSMUST00000177932.7 ENSMUST00000201349.6 |
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr15_-_88863210 | 0.01 |
ENSMUST00000042594.13
ENSMUST00000109368.2 |
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) |
| chrX_+_9066105 | 0.01 |
ENSMUST00000069763.3
|
Lancl3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chr1_+_21419819 | 0.01 |
ENSMUST00000088407.4
|
Khdc1a
|
KH domain containing 1A |
| chrX_+_72235828 | 0.01 |
ENSMUST00000101486.5
|
Xlr3b
|
X-linked lymphocyte-regulated 3B |
| chr7_+_12246415 | 0.01 |
ENSMUST00000032541.5
|
2900092C05Rik
|
RIKEN cDNA 2900092C05 gene |
| chr16_-_13548833 | 0.01 |
ENSMUST00000023364.7
|
Pla2g10
|
phospholipase A2, group X |
| chr5_-_121641461 | 0.01 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr3_-_87985602 | 0.01 |
ENSMUST00000050258.9
|
Ttc24
|
tetratricopeptide repeat domain 24 |
| chr16_-_13548307 | 0.01 |
ENSMUST00000115807.9
|
Pla2g10
|
phospholipase A2, group X |
| chr7_+_80764547 | 0.01 |
ENSMUST00000026820.11
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr14_-_56397036 | 0.01 |
ENSMUST00000015578.5
|
Gzmg
|
granzyme G |
| chr5_-_103247920 | 0.01 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr14_+_9646630 | 0.00 |
ENSMUST00000112658.8
ENSMUST00000112657.9 ENSMUST00000177814.2 ENSMUST00000067491.14 |
Cadps
|
Ca2+-dependent secretion activator |
| chr1_-_74974707 | 0.00 |
ENSMUST00000094844.4
|
Cfap65
|
cilia and flagella associated protein 65 |
| chr11_+_117375194 | 0.00 |
ENSMUST00000092394.4
|
Gm11733
|
predicted gene 11733 |
| chr15_+_80862074 | 0.00 |
ENSMUST00000229727.2
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr2_-_111476340 | 0.00 |
ENSMUST00000119566.4
|
Olfr1298
|
olfactory receptor 1298 |
| chr1_-_65071897 | 0.00 |
ENSMUST00000162800.8
ENSMUST00000069142.12 |
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr7_-_64575308 | 0.00 |
ENSMUST00000149851.8
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr9_+_36743980 | 0.00 |
ENSMUST00000034630.15
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr3_+_99147677 | 0.00 |
ENSMUST00000151606.8
|
Tbx15
|
T-box 15 |
| chr5_+_95604665 | 0.00 |
ENSMUST00000094593.7
|
Pramel48
|
PRAME like 48 |
| chr15_-_79048674 | 0.00 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr11_+_49039086 | 0.00 |
ENSMUST00000059379.2
|
Olfr1395
|
olfactory receptor 1395 |
| chr10_+_119655294 | 0.00 |
ENSMUST00000105262.9
ENSMUST00000147454.8 ENSMUST00000138410.8 ENSMUST00000144825.8 ENSMUST00000148954.8 ENSMUST00000144959.8 |
Grip1
|
glutamate receptor interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.3 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.3 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.6 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.3 | GO:0070469 | respiratory chain(GO:0070469) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.2 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 1.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |