Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
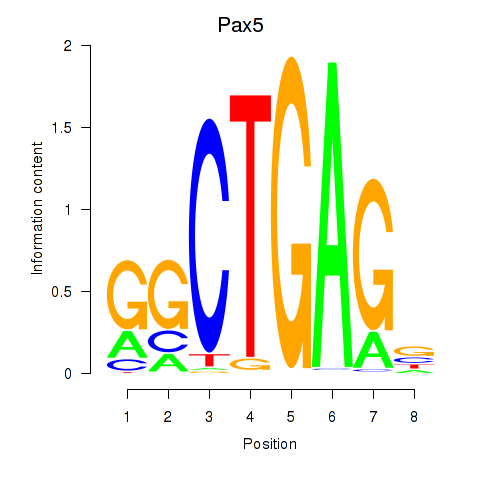
Results for Pax5
Z-value: 1.55
Transcription factors associated with Pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax5
|
ENSMUSG00000014030.16 | paired box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax5 | mm39_v1_chr4_-_44710408_44710487 | 0.94 | 1.6e-02 | Click! |
Activity profile of Pax5 motif
Sorted Z-values of Pax5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_51045182 | 1.45 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr7_-_102148780 | 0.74 |
ENSMUST00000216116.4
|
Olfr545
|
olfactory receptor 545 |
| chr15_+_6599001 | 0.72 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr2_+_165834546 | 0.70 |
ENSMUST00000109252.8
ENSMUST00000088095.6 |
Ncoa3
|
nuclear receptor coactivator 3 |
| chr5_-_116427003 | 0.69 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr15_-_82783978 | 0.69 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr15_+_102885467 | 0.67 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr10_+_127612243 | 0.63 |
ENSMUST00000136223.2
ENSMUST00000052652.7 |
Rdh9
|
retinol dehydrogenase 9 |
| chr10_-_10958031 | 0.62 |
ENSMUST00000105561.9
ENSMUST00000044306.13 |
Grm1
|
glutamate receptor, metabotropic 1 |
| chr10_-_7831657 | 0.62 |
ENSMUST00000147938.2
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr4_-_41569500 | 0.60 |
ENSMUST00000108049.9
ENSMUST00000108052.10 ENSMUST00000108050.2 |
Fam219a
|
family with sequence similarity 219, member A |
| chr2_-_130748259 | 0.59 |
ENSMUST00000128176.2
ENSMUST00000133766.2 ENSMUST00000135110.2 ENSMUST00000146975.2 |
A730017L22Rik
4930402H24Rik
|
RIKEN cDNA A730017L22 gene RIKEN cDNA 4930402H24 gene |
| chr2_-_104324035 | 0.59 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr10_+_127595590 | 0.57 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr11_+_68986043 | 0.57 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr15_+_101191099 | 0.56 |
ENSMUST00000191426.2
|
Smim41
|
small integral membrane protein 41 |
| chr2_-_24365607 | 0.56 |
ENSMUST00000028355.11
|
Pax8
|
paired box 8 |
| chr9_+_118892497 | 0.54 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr2_-_24365576 | 0.51 |
ENSMUST00000153601.8
ENSMUST00000136228.10 |
Pax8
|
paired box 8 |
| chr17_+_27171834 | 0.50 |
ENSMUST00000231853.2
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr19_+_8966641 | 0.50 |
ENSMUST00000092956.4
ENSMUST00000092955.11 |
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr14_+_20732804 | 0.50 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr13_+_58956495 | 0.49 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr13_+_58956077 | 0.48 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr7_-_97958679 | 0.48 |
ENSMUST00000033020.14
|
Acer3
|
alkaline ceramidase 3 |
| chr12_+_84616536 | 0.47 |
ENSMUST00000021665.12
ENSMUST00000169934.4 |
Vsx2
|
visual system homeobox 2 |
| chr15_+_81469538 | 0.47 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr7_-_117842892 | 0.47 |
ENSMUST00000179047.3
|
Smg1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
| chr15_+_6609322 | 0.46 |
ENSMUST00000090461.12
|
Fyb
|
FYN binding protein |
| chr17_+_35455532 | 0.46 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr11_+_98754434 | 0.45 |
ENSMUST00000142414.8
ENSMUST00000037480.9 |
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr18_-_60724855 | 0.45 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr1_+_37258272 | 0.44 |
ENSMUST00000027288.10
|
Cnga3
|
cyclic nucleotide gated channel alpha 3 |
| chr7_+_3339077 | 0.44 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr17_+_84013575 | 0.44 |
ENSMUST00000112350.8
ENSMUST00000112349.9 ENSMUST00000112352.10 ENSMUST00000067826.15 |
Mta3
|
metastasis associated 3 |
| chr7_-_19338349 | 0.44 |
ENSMUST00000086041.7
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr4_-_156312961 | 0.43 |
ENSMUST00000217885.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr11_+_80044931 | 0.43 |
ENSMUST00000021050.14
|
Adap2
|
ArfGAP with dual PH domains 2 |
| chr7_+_3339059 | 0.42 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr9_-_110305705 | 0.42 |
ENSMUST00000198164.5
ENSMUST00000068025.13 |
Klhl18
|
kelch-like 18 |
| chr19_+_3373285 | 0.42 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr4_+_119590971 | 0.41 |
ENSMUST00000084306.5
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr1_-_72914036 | 0.41 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr17_+_35235552 | 0.41 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr7_+_27878894 | 0.40 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr1_-_60605867 | 0.40 |
ENSMUST00000027168.12
ENSMUST00000090293.11 ENSMUST00000140485.8 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr14_+_115329676 | 0.39 |
ENSMUST00000176912.8
ENSMUST00000175665.8 |
Gpc5
|
glypican 5 |
| chr1_+_62742444 | 0.38 |
ENSMUST00000102822.9
ENSMUST00000075144.12 |
Nrp2
|
neuropilin 2 |
| chr2_-_70885877 | 0.38 |
ENSMUST00000090849.6
ENSMUST00000100037.9 ENSMUST00000112186.9 |
Mettl8
|
methyltransferase like 8 |
| chr8_-_106198112 | 0.38 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr11_+_83637766 | 0.37 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chr4_+_150699670 | 0.37 |
ENSMUST00000219467.2
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr6_-_35110560 | 0.37 |
ENSMUST00000202143.4
ENSMUST00000114993.9 ENSMUST00000114989.9 ENSMUST00000044163.10 ENSMUST00000202417.2 |
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr17_-_16046780 | 0.37 |
ENSMUST00000232638.2
ENSMUST00000170578.3 |
Rgmb
|
repulsive guidance molecule family member B |
| chr16_+_14523696 | 0.37 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr7_+_26895206 | 0.36 |
ENSMUST00000179391.8
ENSMUST00000108379.8 |
BC024978
|
cDNA sequence BC024978 |
| chr7_+_127078371 | 0.36 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr6_-_38852899 | 0.35 |
ENSMUST00000160360.2
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr4_-_156312996 | 0.35 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr16_+_24266829 | 0.34 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_101151394 | 0.34 |
ENSMUST00000103108.8
|
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr6_-_38852857 | 0.33 |
ENSMUST00000162359.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr2_+_113271409 | 0.33 |
ENSMUST00000081349.9
|
Fmn1
|
formin 1 |
| chr13_-_24118139 | 0.33 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr11_-_4110286 | 0.33 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr14_-_105414681 | 0.32 |
ENSMUST00000163499.2
|
Rbm26
|
RNA binding motif protein 26 |
| chr10_-_9550907 | 0.32 |
ENSMUST00000100070.5
|
Samd5
|
sterile alpha motif domain containing 5 |
| chr3_+_40848580 | 0.32 |
ENSMUST00000159774.7
ENSMUST00000203472.3 ENSMUST00000203650.3 ENSMUST00000108077.10 ENSMUST00000203892.2 |
Abhd18
|
abhydrolase domain containing 18 |
| chr9_-_91247809 | 0.32 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr1_+_173093568 | 0.32 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr4_+_73931679 | 0.32 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr7_+_75259778 | 0.32 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr12_+_33364288 | 0.31 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr2_-_155676765 | 0.31 |
ENSMUST00000029143.7
ENSMUST00000239423.2 |
Fam83c
|
family with sequence similarity 83, member C |
| chr17_+_24022153 | 0.31 |
ENSMUST00000190686.7
ENSMUST00000088621.11 ENSMUST00000233636.2 |
Srrm2
|
serine/arginine repetitive matrix 2 |
| chrX_+_20554193 | 0.31 |
ENSMUST00000115364.8
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr11_+_66802807 | 0.31 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr7_+_96730915 | 0.30 |
ENSMUST00000206791.2
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr18_-_67743854 | 0.30 |
ENSMUST00000115050.10
|
Spire1
|
spire type actin nucleation factor 1 |
| chr1_-_139304779 | 0.30 |
ENSMUST00000196402.5
ENSMUST00000059825.12 |
Crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr18_+_36481706 | 0.30 |
ENSMUST00000235864.2
ENSMUST00000050584.10 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr2_-_89951611 | 0.30 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr11_+_54413772 | 0.29 |
ENSMUST00000207429.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chrX_-_101114906 | 0.29 |
ENSMUST00000188731.2
|
Rtl5
|
retrotransposon Gag like 5 |
| chr11_-_119989492 | 0.29 |
ENSMUST00000093901.12
ENSMUST00000026442.11 ENSMUST00000106225.4 |
Tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr14_-_21102487 | 0.29 |
ENSMUST00000154460.8
ENSMUST00000130291.8 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr7_-_19556612 | 0.29 |
ENSMUST00000120537.8
|
Bcl3
|
B cell leukemia/lymphoma 3 |
| chr7_-_28297565 | 0.29 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chrX_+_102400061 | 0.29 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr1_-_160079007 | 0.29 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr2_+_120459593 | 0.28 |
ENSMUST00000180041.9
|
Stard9
|
START domain containing 9 |
| chr3_+_45332831 | 0.28 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr10_-_49664839 | 0.28 |
ENSMUST00000220263.2
ENSMUST00000218823.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr11_-_76969230 | 0.28 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr2_-_38177359 | 0.28 |
ENSMUST00000102787.10
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr15_-_90563510 | 0.28 |
ENSMUST00000014777.9
ENSMUST00000064391.12 |
Cpne8
|
copine VIII |
| chr18_-_25301729 | 0.28 |
ENSMUST00000148255.8
|
Tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr8_+_85449632 | 0.28 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr4_-_82803384 | 0.28 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr16_-_23339548 | 0.28 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chrX_-_101114794 | 0.28 |
ENSMUST00000113631.2
|
Rtl5
|
retrotransposon Gag like 5 |
| chr11_-_102446947 | 0.28 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8 |
| chrX_+_20554618 | 0.28 |
ENSMUST00000033380.7
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr14_+_54862762 | 0.28 |
ENSMUST00000097177.5
|
Psmb11
|
proteasome (prosome, macropain) subunit, beta type, 11 |
| chr2_-_45000389 | 0.27 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr4_-_123507494 | 0.27 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr4_-_119217079 | 0.27 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr2_+_28082943 | 0.27 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1 |
| chr4_+_132701413 | 0.27 |
ENSMUST00000030693.13
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr16_-_46317318 | 0.27 |
ENSMUST00000023335.13
ENSMUST00000023334.15 |
Nectin3
|
nectin cell adhesion molecule 3 |
| chr4_+_124774692 | 0.27 |
ENSMUST00000059343.7
|
Epha10
|
Eph receptor A10 |
| chr9_+_54888750 | 0.27 |
ENSMUST00000217408.2
ENSMUST00000213960.2 |
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr5_+_129661233 | 0.26 |
ENSMUST00000031390.10
|
Mmp17
|
matrix metallopeptidase 17 |
| chr6_+_68402550 | 0.26 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr2_-_111400026 | 0.26 |
ENSMUST00000217772.2
ENSMUST00000207283.3 |
Olfr1295
|
olfactory receptor 1295 |
| chr3_-_103698391 | 0.26 |
ENSMUST00000106845.9
ENSMUST00000029438.15 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr11_-_115494692 | 0.26 |
ENSMUST00000125097.2
ENSMUST00000019135.14 ENSMUST00000106508.10 |
Gga3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr1_+_34044940 | 0.26 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr14_+_63075127 | 0.26 |
ENSMUST00000014691.10
|
Wdfy2
|
WD repeat and FYVE domain containing 2 |
| chr2_+_79538124 | 0.26 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr17_-_66384017 | 0.26 |
ENSMUST00000150766.2
ENSMUST00000038116.13 |
Ankrd12
|
ankyrin repeat domain 12 |
| chr13_+_95833359 | 0.26 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr3_-_89959739 | 0.26 |
ENSMUST00000199929.2
ENSMUST00000090908.11 ENSMUST00000198322.5 ENSMUST00000196843.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr7_+_131568167 | 0.26 |
ENSMUST00000045840.5
|
Gpr26
|
G protein-coupled receptor 26 |
| chr4_+_83335947 | 0.26 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr19_+_5754395 | 0.26 |
ENSMUST00000052448.4
|
Kcnk7
|
potassium channel, subfamily K, member 7 |
| chrX_+_149330371 | 0.26 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr2_-_162502994 | 0.25 |
ENSMUST00000109442.8
ENSMUST00000109445.9 ENSMUST00000109443.8 ENSMUST00000109441.2 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr13_+_54519161 | 0.25 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr11_+_54413698 | 0.25 |
ENSMUST00000108895.8
ENSMUST00000101206.10 |
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr3_-_75864195 | 0.25 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr11_-_101117237 | 0.25 |
ENSMUST00000100417.3
ENSMUST00000107285.8 ENSMUST00000107284.8 |
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr6_-_124746510 | 0.25 |
ENSMUST00000149652.2
ENSMUST00000112476.8 ENSMUST00000004378.15 |
Eno2
|
enolase 2, gamma neuronal |
| chr14_-_76348179 | 0.25 |
ENSMUST00000022585.5
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr3_-_94693740 | 0.25 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr17_-_46991709 | 0.25 |
ENSMUST00000233524.2
ENSMUST00000233733.2 ENSMUST00000071841.7 ENSMUST00000165007.9 |
Klhdc3
|
kelch domain containing 3 |
| chr9_-_56325344 | 0.25 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr2_+_156562956 | 0.25 |
ENSMUST00000109566.9
ENSMUST00000146412.9 ENSMUST00000177013.8 ENSMUST00000171030.9 |
Dlgap4
|
DLG associated protein 4 |
| chr15_-_101262452 | 0.24 |
ENSMUST00000230909.2
|
Krt80
|
keratin 80 |
| chr2_-_25873068 | 0.24 |
ENSMUST00000127823.2
ENSMUST00000134882.8 |
Camsap1
|
calmodulin regulated spectrin-associated protein 1 |
| chr2_+_31578537 | 0.24 |
ENSMUST00000075759.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr19_-_4213347 | 0.24 |
ENSMUST00000025749.15
|
Rps6kb2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr11_+_77873535 | 0.24 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr14_-_105414714 | 0.24 |
ENSMUST00000100327.10
ENSMUST00000022715.14 |
Rbm26
|
RNA binding motif protein 26 |
| chr12_-_115832846 | 0.24 |
ENSMUST00000199373.2
|
Ighv1-78
|
immunoglobulin heavy variable 1-78 |
| chr7_-_126101555 | 0.24 |
ENSMUST00000167759.8
|
Atxn2l
|
ataxin 2-like |
| chr8_-_106553822 | 0.24 |
ENSMUST00000239468.2
ENSMUST00000041400.6 |
Ranbp10
|
RAN binding protein 10 |
| chr4_-_155870321 | 0.24 |
ENSMUST00000097742.3
|
Tmem88b
|
transmembrane protein 88B |
| chr10_+_59942274 | 0.23 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr8_-_84299540 | 0.23 |
ENSMUST00000212990.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr15_-_81074921 | 0.23 |
ENSMUST00000131235.9
ENSMUST00000134469.9 ENSMUST00000239114.2 ENSMUST00000149582.8 |
Mrtfa
|
myocardin related transcription factor A |
| chr3_-_152687877 | 0.23 |
ENSMUST00000044278.6
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr3_-_89959770 | 0.23 |
ENSMUST00000029553.16
ENSMUST00000195995.5 ENSMUST00000064639.15 ENSMUST00000199834.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr16_+_13074345 | 0.23 |
ENSMUST00000009713.14
ENSMUST00000115809.8 |
Mrtfb
|
myocardin related transcription factor B |
| chr5_-_144294854 | 0.23 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr10_+_107998033 | 0.23 |
ENSMUST00000219263.2
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr17_+_29020744 | 0.23 |
ENSMUST00000136233.2
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr16_-_18904240 | 0.23 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr15_-_79543090 | 0.23 |
ENSMUST00000109648.9
|
Fam227a
|
family with sequence similarity 227, member A |
| chr11_+_70431063 | 0.22 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chrX_+_7439839 | 0.22 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr7_-_126101484 | 0.22 |
ENSMUST00000166682.9
|
Atxn2l
|
ataxin 2-like |
| chr13_-_61084358 | 0.22 |
ENSMUST00000225859.2
ENSMUST00000225167.2 ENSMUST00000021880.10 |
Gm49391
Ctla2a
|
predicted gene, 49391 cytotoxic T lymphocyte-associated protein 2 alpha |
| chr9_-_44145280 | 0.22 |
ENSMUST00000205968.2
ENSMUST00000206147.2 ENSMUST00000037644.8 |
Cbl
|
Casitas B-lineage lymphoma |
| chr7_-_117842787 | 0.22 |
ENSMUST00000032891.15
|
Smg1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
| chr4_-_123558516 | 0.22 |
ENSMUST00000147030.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr7_-_123099672 | 0.22 |
ENSMUST00000042470.14
ENSMUST00000128217.2 |
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr17_+_29077385 | 0.22 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr11_+_77576981 | 0.22 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr17_-_26417982 | 0.22 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr14_-_105414294 | 0.22 |
ENSMUST00000163545.8
|
Rbm26
|
RNA binding motif protein 26 |
| chr15_+_80595486 | 0.22 |
ENSMUST00000067689.9
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr2_+_119727689 | 0.22 |
ENSMUST00000046717.13
ENSMUST00000079934.12 ENSMUST00000110774.8 ENSMUST00000110773.9 ENSMUST00000156510.2 |
Mga
|
MAX gene associated |
| chr11_+_101169918 | 0.22 |
ENSMUST00000103107.5
|
Cntd1
|
cyclin N-terminal domain containing 1 |
| chr9_+_119978773 | 0.22 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr13_+_19807274 | 0.21 |
ENSMUST00000222464.2
ENSMUST00000002883.7 |
Sfrp4
|
secreted frizzled-related protein 4 |
| chr2_+_28083105 | 0.21 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1 |
| chr2_-_45000250 | 0.21 |
ENSMUST00000201211.4
ENSMUST00000177302.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_3165485 | 0.21 |
ENSMUST00000232048.2
|
Scaf8
|
SR-related CTD-associated factor 8 |
| chr11_+_68447012 | 0.21 |
ENSMUST00000053211.8
|
Mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr7_-_19621833 | 0.21 |
ENSMUST00000052605.8
|
Ceacam19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr7_+_6459167 | 0.21 |
ENSMUST00000214301.3
|
Olfr1336
|
olfactory receptor 1336 |
| chr2_-_38177182 | 0.21 |
ENSMUST00000130472.8
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr8_+_71207326 | 0.21 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr12_-_87037204 | 0.21 |
ENSMUST00000222543.2
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr19_-_4384029 | 0.21 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr13_-_19917092 | 0.21 |
ENSMUST00000151029.3
|
Gpr141b
|
G protein-coupled receptor 141B |
| chr11_-_69435382 | 0.21 |
ENSMUST00000035539.12
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr8_-_106016496 | 0.21 |
ENSMUST00000014981.8
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr5_-_92404137 | 0.21 |
ENSMUST00000201130.4
|
Ppef2
|
protein phosphatase, EF hand calcium-binding domain 2 |
| chr9_-_44632680 | 0.21 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr8_+_26092252 | 0.21 |
ENSMUST00000136107.9
ENSMUST00000146919.8 ENSMUST00000139966.8 ENSMUST00000142395.8 ENSMUST00000143445.2 |
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr11_+_54413673 | 0.21 |
ENSMUST00000102743.10
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr11_-_90528888 | 0.21 |
ENSMUST00000020858.14
ENSMUST00000107875.8 ENSMUST00000107872.8 ENSMUST00000143203.8 |
Stxbp4
|
syntaxin binding protein 4 |
| chr17_-_36220924 | 0.20 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr10_+_59942020 | 0.20 |
ENSMUST00000121820.9
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr5_-_25703700 | 0.20 |
ENSMUST00000173073.8
ENSMUST00000045291.14 ENSMUST00000173174.2 |
Kmt2c
|
lysine (K)-specific methyltransferase 2C |
| chr18_+_35695736 | 0.20 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr7_-_46445305 | 0.20 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr3_+_131270648 | 0.20 |
ENSMUST00000199878.5
ENSMUST00000196408.5 ENSMUST00000200527.5 |
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.2 | 0.5 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.2 | 1.0 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.2 | 0.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.3 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.9 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.7 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.1 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.2 | GO:0002851 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 0.3 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.0 | 0.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.2 | GO:0046722 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.4 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.4 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.9 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0014854 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:0061744 | motor behavior(GO:0061744) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.1 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.2 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.4 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 1.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.7 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.5 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 1.0 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.0 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.4 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) |
| 0.0 | 0.6 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:0034088 | maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.0 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.0 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.1 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.0 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 1.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.2 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.2 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.6 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.0 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 1.0 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.4 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:1902121 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |