Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Pax6
Z-value: 1.05
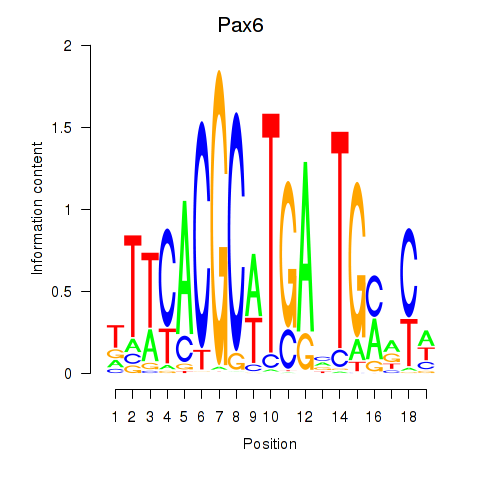
Transcription factors associated with Pax6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax6
|
ENSMUSG00000027168.22 | paired box 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax6 | mm39_v1_chr2_+_105505823_105505872 | 0.69 | 2.0e-01 | Click! |
Activity profile of Pax6 motif
Sorted Z-values of Pax6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_110416637 | 1.40 |
ENSMUST00000167824.3
ENSMUST00000224180.2 |
Rab3c
|
RAB3C, member RAS oncogene family |
| chrX_+_16485937 | 0.93 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr4_+_85123654 | 0.82 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr7_+_5054514 | 0.79 |
ENSMUST00000069324.7
|
Zfp580
|
zinc finger protein 580 |
| chr1_+_59802543 | 0.74 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr8_-_73228953 | 0.69 |
ENSMUST00000079510.6
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr7_+_86645323 | 0.68 |
ENSMUST00000233714.2
ENSMUST00000233648.2 ENSMUST00000164462.3 ENSMUST00000233730.2 |
Vmn2r79
|
vomeronasal 2, receptor 79 |
| chr8_-_73229056 | 0.63 |
ENSMUST00000212991.2
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr2_+_36795692 | 0.59 |
ENSMUST00000217479.2
|
Olfr354
|
olfactory receptor 354 |
| chr1_+_131566044 | 0.57 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr2_+_111084861 | 0.57 |
ENSMUST00000218065.2
|
Olfr1276
|
olfactory receptor 1276 |
| chr4_-_123507494 | 0.57 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr7_+_75259778 | 0.49 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr8_+_110341049 | 0.48 |
ENSMUST00000057344.3
ENSMUST00000212537.2 ENSMUST00000109242.8 |
Pkd1l3
|
polycystic kidney disease 1 like 3 |
| chr17_-_57366795 | 0.43 |
ENSMUST00000040280.14
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr2_-_111400026 | 0.41 |
ENSMUST00000217772.2
ENSMUST00000207283.3 |
Olfr1295
|
olfactory receptor 1295 |
| chr15_-_35938328 | 0.38 |
ENSMUST00000014457.15
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr7_+_97102411 | 0.37 |
ENSMUST00000121987.3
ENSMUST00000050732.14 ENSMUST00000205577.2 ENSMUST00000206279.2 |
Kctd14
|
potassium channel tetramerisation domain containing 14 |
| chr6_-_122833109 | 0.37 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr10_+_129320621 | 0.36 |
ENSMUST00000213236.2
ENSMUST00000213992.2 |
Olfr789
|
olfactory receptor 789 |
| chr4_+_148123554 | 0.34 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr11_-_104441218 | 0.34 |
ENSMUST00000106962.9
ENSMUST00000106961.2 ENSMUST00000093923.9 |
Cdc27
|
cell division cycle 27 |
| chr2_-_111104451 | 0.33 |
ENSMUST00000214760.2
|
Olfr1277
|
olfactory receptor 1277 |
| chr12_+_76593799 | 0.32 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr11_+_33996920 | 0.32 |
ENSMUST00000052413.12
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chrX_+_23559282 | 0.29 |
ENSMUST00000035766.13
ENSMUST00000101670.3 |
Wdr44
|
WD repeat domain 44 |
| chr18_+_37580692 | 0.25 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr11_-_95478517 | 0.25 |
ENSMUST00000000122.7
|
Ngfr
|
nerve growth factor receptor (TNFR superfamily, member 16) |
| chr13_-_110417421 | 0.23 |
ENSMUST00000223922.2
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr7_+_140808680 | 0.23 |
ENSMUST00000106027.9
|
Phrf1
|
PHD and ring finger domains 1 |
| chr9_+_64086553 | 0.21 |
ENSMUST00000034965.8
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr6_-_88021999 | 0.21 |
ENSMUST00000113598.8
|
Rab7
|
RAB7, member RAS oncogene family |
| chr16_+_3702523 | 0.21 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr19_+_13385213 | 0.19 |
ENSMUST00000216910.3
|
Olfr1469
|
olfactory receptor 1469 |
| chr7_-_101517874 | 0.18 |
ENSMUST00000150184.2
|
Folr1
|
folate receptor 1 (adult) |
| chr11_+_78219241 | 0.17 |
ENSMUST00000048073.9
|
Pigs
|
phosphatidylinositol glycan anchor biosynthesis, class S |
| chr6_-_113354337 | 0.13 |
ENSMUST00000043333.9
|
Tada3
|
transcriptional adaptor 3 |
| chr11_+_69016722 | 0.13 |
ENSMUST00000021268.9
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr7_+_86444235 | 0.12 |
ENSMUST00000233099.2
ENSMUST00000164996.2 |
Vmn2r77
|
vomeronasal 2, receptor 77 |
| chr2_-_33777874 | 0.12 |
ENSMUST00000041555.10
|
Mvb12b
|
multivesicular body subunit 12B |
| chr8_+_106587212 | 0.11 |
ENSMUST00000008594.9
|
Nutf2
|
nuclear transport factor 2 |
| chr6_-_144994534 | 0.11 |
ENSMUST00000032402.12
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr4_+_138920210 | 0.11 |
ENSMUST00000102508.10
ENSMUST00000131912.8 ENSMUST00000102507.10 |
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr6_+_29468067 | 0.11 |
ENSMUST00000143101.4
ENSMUST00000149646.3 |
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr2_+_37029334 | 0.11 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chr11_+_72981377 | 0.09 |
ENSMUST00000006101.4
|
Itgae
|
integrin alpha E, epithelial-associated |
| chr2_-_65068917 | 0.08 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr11_+_67857268 | 0.08 |
ENSMUST00000021286.11
ENSMUST00000108675.2 |
Stx8
|
syntaxin 8 |
| chr11_-_115503316 | 0.07 |
ENSMUST00000106507.9
|
Mif4gd
|
MIF4G domain containing |
| chr16_+_34605282 | 0.07 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr11_-_72097821 | 0.07 |
ENSMUST00000204457.3
|
Gm43951
|
predicted gene, 43951 |
| chr15_+_95698574 | 0.07 |
ENSMUST00000226793.2
|
Ano6
|
anoctamin 6 |
| chr8_-_81607140 | 0.07 |
ENSMUST00000210676.2
|
Gab1
|
growth factor receptor bound protein 2-associated protein 1 |
| chr2_-_117173312 | 0.06 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr6_-_125213911 | 0.06 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr4_-_140787852 | 0.06 |
ENSMUST00000144196.2
ENSMUST00000097816.9 |
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr13_-_113755082 | 0.06 |
ENSMUST00000109241.5
|
Snx18
|
sorting nexin 18 |
| chr11_-_121410152 | 0.05 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr13_+_63963054 | 0.05 |
ENSMUST00000021926.13
ENSMUST00000067821.13 ENSMUST00000144763.2 ENSMUST00000021925.14 ENSMUST00000238465.2 |
Ercc6l2
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 like 2 |
| chr16_+_3702604 | 0.04 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr19_+_34268071 | 0.04 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr17_-_43854530 | 0.04 |
ENSMUST00000178772.3
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr4_-_70328659 | 0.03 |
ENSMUST00000144099.8
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chrX_+_73784533 | 0.03 |
ENSMUST00000114117.2
|
Ctag2l2
|
CTAG2 like 2 |
| chr19_+_34560922 | 0.03 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr7_+_51160754 | 0.02 |
ENSMUST00000043944.6
ENSMUST00000207044.2 |
Ano5
|
anoctamin 5 |
| chr6_+_60921456 | 0.02 |
ENSMUST00000129603.4
ENSMUST00000204333.2 |
Mmrn1
|
multimerin 1 |
| chr1_-_174079627 | 0.02 |
ENSMUST00000214725.2
|
Olfr419
|
olfactory receptor 419 |
| chr11_+_72326337 | 0.02 |
ENSMUST00000076443.10
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr17_-_33937565 | 0.02 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr17_+_25585255 | 0.02 |
ENSMUST00000234477.2
|
Tpsb2
|
tryptase beta 2 |
| chr10_-_127025851 | 0.02 |
ENSMUST00000222006.2
ENSMUST00000019611.15 ENSMUST00000219245.2 |
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr5_+_110248276 | 0.01 |
ENSMUST00000141066.8
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr1_+_21310821 | 0.01 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_63215976 | 0.01 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr9_+_105520154 | 0.01 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr10_+_80192293 | 0.01 |
ENSMUST00000039836.15
ENSMUST00000105351.2 |
Plk5
|
polo like kinase 5 |
| chr11_-_67856457 | 0.01 |
ENSMUST00000021287.12
ENSMUST00000126766.2 |
Cfap52
|
cilia and flagella associated protein 52 |
| chr7_-_126408280 | 0.01 |
ENSMUST00000207534.3
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr8_-_81607109 | 0.01 |
ENSMUST00000034150.10
|
Gab1
|
growth factor receptor bound protein 2-associated protein 1 |
| chrX_-_111316476 | 0.01 |
ENSMUST00000026601.3
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr2_+_120331784 | 0.01 |
ENSMUST00000151342.3
|
Capn3
|
calpain 3 |
| chr11_+_72326358 | 0.01 |
ENSMUST00000108499.2
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr11_+_104441489 | 0.01 |
ENSMUST00000018800.9
|
Myl4
|
myosin, light polypeptide 4 |
| chrX_-_73525005 | 0.01 |
ENSMUST00000114125.2
|
Ctag2l1
|
CTAG2 like 1 |
| chr6_-_144994327 | 0.01 |
ENSMUST00000204138.3
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr12_-_11200306 | 0.01 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr7_+_63803663 | 0.01 |
ENSMUST00000206314.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr7_+_51160855 | 0.01 |
ENSMUST00000207717.2
|
Ano5
|
anoctamin 5 |
| chr4_-_139560229 | 0.01 |
ENSMUST00000174681.2
|
Pax7
|
paired box 7 |
| chr9_-_49338321 | 0.00 |
ENSMUST00000034792.7
|
Ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr7_-_103492361 | 0.00 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr5_+_142615292 | 0.00 |
ENSMUST00000036872.16
ENSMUST00000110778.2 |
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr9_+_15150341 | 0.00 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr13_-_4573312 | 0.00 |
ENSMUST00000221564.2
ENSMUST00000078239.5 ENSMUST00000080361.13 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr9_-_54554483 | 0.00 |
ENSMUST00000128163.8
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr4_-_71676202 | 0.00 |
ENSMUST00000084489.3
|
Gm11232
|
predicted gene 11232 |
| chr11_+_72326391 | 0.00 |
ENSMUST00000100903.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr9_-_117701613 | 0.00 |
ENSMUST00000239475.2
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr8_-_27718522 | 0.00 |
ENSMUST00000117565.2
|
Adrb3
|
adrenergic receptor, beta 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 1.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.4 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.1 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.2 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.1 | 0.4 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.8 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 1.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.0 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) alveolar lamellar body(GO:0097208) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.7 | GO:0098821 | BMP binding(GO:0036122) BMP receptor activity(GO:0098821) |
| 0.1 | 1.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |