Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
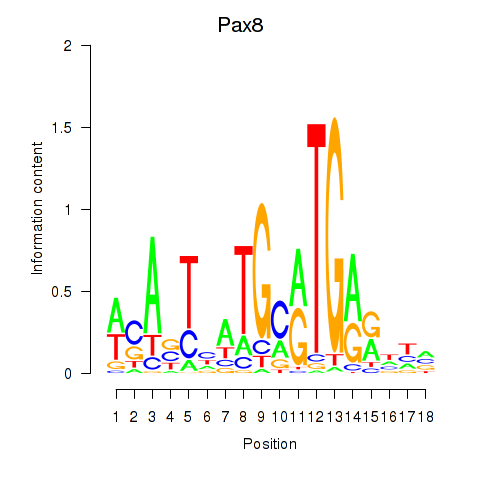
Results for Pax8
Z-value: 0.92
Transcription factors associated with Pax8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax8
|
ENSMUSG00000026976.16 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax8 | mm39_v1_chr2_-_24365607_24365619 | 1.00 | 3.9e-05 | Click! |
Activity profile of Pax8 motif
Sorted Z-values of Pax8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_50683002 | 1.20 |
ENSMUST00000214792.2
|
Olfr740
|
olfactory receptor 740 |
| chr5_+_107655487 | 1.05 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr7_-_30560989 | 0.76 |
ENSMUST00000052700.6
|
Ffar1
|
free fatty acid receptor 1 |
| chr12_+_85043262 | 0.52 |
ENSMUST00000101202.10
|
Ylpm1
|
YLP motif containing 1 |
| chr1_+_165591315 | 0.46 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr12_+_85043083 | 0.43 |
ENSMUST00000168977.8
ENSMUST00000021670.15 |
Ylpm1
|
YLP motif containing 1 |
| chr14_+_50741057 | 0.39 |
ENSMUST00000217437.2
ENSMUST00000213935.2 |
Olfr742
|
olfactory receptor 742 |
| chr12_+_71095112 | 0.33 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr3_+_138058139 | 0.32 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr11_-_69692542 | 0.31 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr3_-_64417263 | 0.30 |
ENSMUST00000177184.9
|
Vmn2r5
|
vomeronasal 2, receptor 5 |
| chr2_+_128809268 | 0.26 |
ENSMUST00000110320.9
ENSMUST00000110319.3 |
Zc3h6
|
zinc finger CCCH type containing 6 |
| chr13_+_108350923 | 0.20 |
ENSMUST00000022207.10
|
Elovl7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| chr1_+_74582044 | 0.14 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr14_+_60615128 | 0.13 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr16_+_34605282 | 0.12 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr15_+_99600475 | 0.12 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr11_+_24030663 | 0.12 |
ENSMUST00000118955.2
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr14_+_50876845 | 0.10 |
ENSMUST00000213127.2
|
Olfr745
|
olfactory receptor 745 |
| chr18_+_69654900 | 0.10 |
ENSMUST00000202057.4
|
Tcf4
|
transcription factor 4 |
| chr1_-_192880260 | 0.10 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr4_+_119397710 | 0.08 |
ENSMUST00000160219.2
|
Foxj3
|
forkhead box J3 |
| chr17_+_45866618 | 0.08 |
ENSMUST00000024742.9
ENSMUST00000233929.2 |
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr19_+_46064409 | 0.08 |
ENSMUST00000223728.2
ENSMUST00000235620.2 |
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr4_+_6191084 | 0.07 |
ENSMUST00000029907.6
|
Ubxn2b
|
UBX domain protein 2B |
| chr1_+_65225886 | 0.07 |
ENSMUST00000097707.5
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr2_-_25471703 | 0.07 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr19_+_46064302 | 0.05 |
ENSMUST00000165017.2
ENSMUST00000223741.2 ENSMUST00000225780.2 |
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr4_+_156300325 | 0.04 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr13_-_62924903 | 0.04 |
ENSMUST00000167516.3
|
Gm5141
|
predicted gene 5141 |
| chr11_-_87788066 | 0.04 |
ENSMUST00000217095.2
ENSMUST00000215150.2 |
Olfr463
|
olfactory receptor 463 |
| chr11_+_78389913 | 0.04 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr6_+_127049865 | 0.04 |
ENSMUST00000000186.9
|
Fgf23
|
fibroblast growth factor 23 |
| chr13_-_63712140 | 0.04 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chrX_+_81992467 | 0.03 |
ENSMUST00000114000.8
|
Dmd
|
dystrophin, muscular dystrophy |
| chr4_-_43429077 | 0.03 |
ENSMUST00000107929.10
ENSMUST00000107928.9 ENSMUST00000171134.9 ENSMUST00000052829.10 |
Fam166b
|
family with sequence similarity 166, member B |
| chr19_+_46064437 | 0.03 |
ENSMUST00000223683.2
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr18_+_69654992 | 0.02 |
ENSMUST00000201627.4
|
Tcf4
|
transcription factor 4 |
| chr14_+_33675934 | 0.01 |
ENSMUST00000035695.10
|
Rbp3
|
retinol binding protein 3, interstitial |
| chr5_+_91222470 | 0.01 |
ENSMUST00000031324.6
|
Ereg
|
epiregulin |
| chr14_+_53220658 | 0.01 |
ENSMUST00000200548.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr4_-_57301475 | 0.01 |
ENSMUST00000130900.2
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr1_-_136877277 | 0.01 |
ENSMUST00000168126.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr2_+_85822163 | 0.01 |
ENSMUST00000050942.3
|
Olfr1031
|
olfactory receptor 1031 |
| chr18_-_60743169 | 0.00 |
ENSMUST00000115318.4
|
Synpo
|
synaptopodin |
| chr18_-_66993567 | 0.00 |
ENSMUST00000057942.4
|
Mc4r
|
melanocortin 4 receptor |
| chr7_-_29605504 | 0.00 |
ENSMUST00000053521.15
|
Zfp27
|
zinc finger protein 27 |
| chr11_-_105346120 | 0.00 |
ENSMUST00000138977.8
|
Marchf10
|
membrane associated ring-CH-type finger 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |