Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
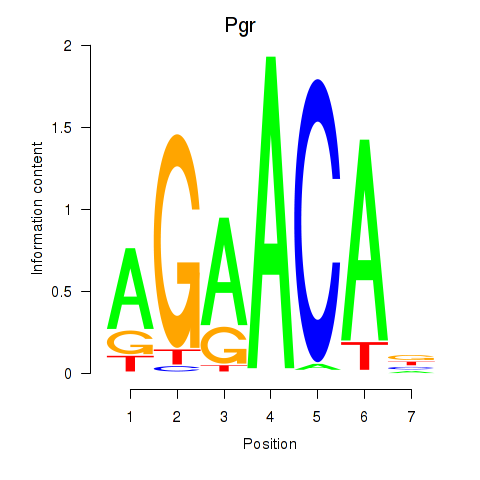
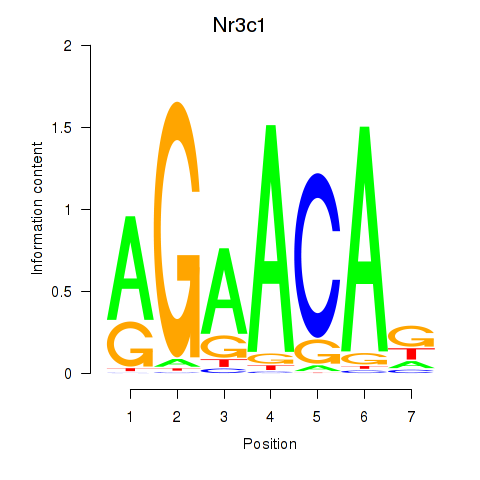
Results for Pgr_Nr3c1
Z-value: 1.28
Transcription factors associated with Pgr_Nr3c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pgr
|
ENSMUSG00000031870.17 | progesterone receptor |
|
Nr3c1
|
ENSMUSG00000024431.16 | nuclear receptor subfamily 3, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c1 | mm39_v1_chr18_-_39652468_39652478 | -0.89 | 4.6e-02 | Click! |
| Pgr | mm39_v1_chr9_+_8899829_8899857 | 0.85 | 7.0e-02 | Click! |
Activity profile of Pgr_Nr3c1 motif
Sorted Z-values of Pgr_Nr3c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_71692320 | 3.11 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr7_+_75259778 | 0.94 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr9_-_49710058 | 0.84 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr6_+_136509922 | 0.77 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_-_66238313 | 0.75 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr11_-_95966477 | 0.73 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr10_-_67972401 | 0.71 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr14_+_99536111 | 0.69 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr9_-_49710190 | 0.68 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr5_-_135494775 | 0.61 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr11_-_3881995 | 0.59 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr5_+_145063568 | 0.57 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr11_-_3881789 | 0.54 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr11_-_3881960 | 0.53 |
ENSMUST00000109990.8
|
Tcn2
|
transcobalamin 2 |
| chr6_+_125297596 | 0.47 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr19_-_6134703 | 0.47 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr1_-_143879738 | 0.45 |
ENSMUST00000153527.3
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr3_-_133250129 | 0.42 |
ENSMUST00000196398.5
ENSMUST00000098603.8 |
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr16_+_51852435 | 0.42 |
ENSMUST00000227879.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr11_+_29413734 | 0.41 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr6_-_87312743 | 0.39 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr15_-_81284244 | 0.38 |
ENSMUST00000172107.8
ENSMUST00000169204.2 ENSMUST00000163382.2 |
St13
|
suppression of tumorigenicity 13 |
| chr4_+_152410291 | 0.34 |
ENSMUST00000103191.11
ENSMUST00000139685.8 ENSMUST00000188151.2 |
Rpl22
|
ribosomal protein L22 |
| chr7_+_28834391 | 0.31 |
ENSMUST00000160194.8
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr14_+_76082736 | 0.30 |
ENSMUST00000142061.3
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr9_+_72832904 | 0.28 |
ENSMUST00000038489.6
|
Pygo1
|
pygopus 1 |
| chr13_+_58956495 | 0.28 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr8_+_66070661 | 0.28 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr11_-_69649452 | 0.28 |
ENSMUST00000058470.16
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr19_-_6134903 | 0.24 |
ENSMUST00000160977.8
ENSMUST00000159859.2 ENSMUST00000025707.9 ENSMUST00000160712.8 ENSMUST00000237738.2 |
Zfpl1
|
zinc finger like protein 1 |
| chr7_+_30252687 | 0.24 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr1_+_88066086 | 0.22 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr4_+_150699670 | 0.21 |
ENSMUST00000219467.2
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr3_-_57599956 | 0.21 |
ENSMUST00000238789.2
ENSMUST00000197088.5 ENSMUST00000099091.4 |
Ankub1
|
ankrin repeat and ubiquitin domain containing 1 |
| chr2_-_153079828 | 0.20 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr8_+_47192767 | 0.20 |
ENSMUST00000034041.9
ENSMUST00000208507.2 ENSMUST00000207105.2 |
Irf2
|
interferon regulatory factor 2 |
| chr4_+_134879807 | 0.20 |
ENSMUST00000119564.2
|
Runx3
|
runt related transcription factor 3 |
| chr8_+_70495417 | 0.20 |
ENSMUST00000011450.8
|
Sugp1
|
SURP and G patch domain containing 1 |
| chr1_-_143879877 | 0.19 |
ENSMUST00000127206.8
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr16_+_43067641 | 0.19 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_53159885 | 0.19 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chrX_-_166113457 | 0.18 |
ENSMUST00000145284.8
ENSMUST00000112164.2 ENSMUST00000137492.8 ENSMUST00000112161.8 ENSMUST00000060719.12 |
Tlr7
|
toll-like receptor 7 |
| chr5_+_30971915 | 0.18 |
ENSMUST00000031058.15
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr5_-_100521343 | 0.18 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr6_-_87312681 | 0.17 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr17_+_36179273 | 0.16 |
ENSMUST00000190496.2
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr15_-_50753792 | 0.15 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr9_+_108356935 | 0.14 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr16_+_20492861 | 0.14 |
ENSMUST00000151679.3
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr15_+_12117899 | 0.13 |
ENSMUST00000122941.8
|
Zfr
|
zinc finger RNA binding protein |
| chr7_-_109215960 | 0.13 |
ENSMUST00000077909.9
|
Denn2b
|
DENN domain containing 2B |
| chr16_+_43184191 | 0.13 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_-_7228555 | 0.13 |
ENSMUST00000063683.8
|
Tagap1
|
T cell activation GTPase activating protein 1 |
| chr19_+_55304703 | 0.12 |
ENSMUST00000225529.2
ENSMUST00000223690.2 ENSMUST00000095950.3 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr1_-_139304779 | 0.12 |
ENSMUST00000196402.5
ENSMUST00000059825.12 |
Crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr6_+_125298296 | 0.12 |
ENSMUST00000081440.14
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr14_-_54949596 | 0.11 |
ENSMUST00000064290.8
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr1_+_34044940 | 0.11 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr11_+_83599841 | 0.11 |
ENSMUST00000001009.14
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr5_+_30972067 | 0.10 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chrX_+_158038778 | 0.10 |
ENSMUST00000126686.8
ENSMUST00000033671.13 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr11_-_23845207 | 0.10 |
ENSMUST00000102863.3
ENSMUST00000020513.10 |
Papolg
|
poly(A) polymerase gamma |
| chr2_-_76503382 | 0.10 |
ENSMUST00000002809.14
|
Fkbp7
|
FK506 binding protein 7 |
| chr12_-_31763859 | 0.09 |
ENSMUST00000057783.6
ENSMUST00000236002.2 ENSMUST00000174480.3 ENSMUST00000176710.2 |
Gpr22
|
G protein-coupled receptor 22 |
| chr5_+_24569802 | 0.09 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr8_+_125302843 | 0.09 |
ENSMUST00000093033.6
ENSMUST00000133086.2 |
Capn9
|
calpain 9 |
| chr6_-_146855880 | 0.08 |
ENSMUST00000111622.2
ENSMUST00000036592.15 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr1_-_57011595 | 0.08 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr16_+_93526987 | 0.08 |
ENSMUST00000227156.2
|
Dop1b
|
DOP1 leucine zipper like protein B |
| chr16_+_38405718 | 0.07 |
ENSMUST00000165631.2
|
Tmem39a
|
transmembrane protein 39a |
| chr3_-_97775557 | 0.07 |
ENSMUST00000107038.6
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr7_-_126184935 | 0.07 |
ENSMUST00000084589.11
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr4_+_119397710 | 0.07 |
ENSMUST00000160219.2
|
Foxj3
|
forkhead box J3 |
| chr2_-_125565307 | 0.07 |
ENSMUST00000042246.14
|
Shc4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr12_-_75782406 | 0.07 |
ENSMUST00000220285.2
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr11_+_117223161 | 0.07 |
ENSMUST00000106349.2
|
Septin9
|
septin 9 |
| chr2_-_76812799 | 0.07 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chrX_+_158038915 | 0.07 |
ENSMUST00000112492.8
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr2_+_79538124 | 0.06 |
ENSMUST00000090760.9
ENSMUST00000040863.11 ENSMUST00000111780.3 |
Ppp1r1c
|
protein phosphatase 1, regulatory inhibitor subunit 1C |
| chr4_+_134042423 | 0.06 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr6_+_42222841 | 0.06 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr2_-_77000936 | 0.06 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr2_+_14179324 | 0.06 |
ENSMUST00000077517.9
|
Tmem236
|
transmembrane protein 236 |
| chr2_-_79959178 | 0.05 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_+_44866635 | 0.05 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr2_-_93826699 | 0.05 |
ENSMUST00000183110.2
|
Gm27027
|
predicted gene, 27027 |
| chr6_+_21949570 | 0.05 |
ENSMUST00000031680.10
ENSMUST00000115389.8 ENSMUST00000151473.8 |
Ing3
|
inhibitor of growth family, member 3 |
| chr15_+_25843225 | 0.05 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr2_-_63014514 | 0.04 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr11_-_110058899 | 0.04 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr6_+_17491231 | 0.04 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr11_+_117716711 | 0.04 |
ENSMUST00000073388.13
|
Afmid
|
arylformamidase |
| chr8_-_70495335 | 0.04 |
ENSMUST00000168013.3
ENSMUST00000212308.2 ENSMUST00000050561.13 ENSMUST00000212451.2 |
Mau2
|
MAU2 sister chromatid cohesion factor |
| chr15_-_98796373 | 0.04 |
ENSMUST00000229775.2
ENSMUST00000023737.6 |
Dhh
|
desert hedgehog |
| chr13_+_91609169 | 0.03 |
ENSMUST00000004094.15
ENSMUST00000042122.15 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr7_-_109215754 | 0.03 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr2_-_65397850 | 0.03 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr14_+_30438334 | 0.03 |
ENSMUST00000228006.2
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr3_-_95811993 | 0.03 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chr14_-_124914516 | 0.03 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr4_-_141125885 | 0.03 |
ENSMUST00000133676.3
ENSMUST00000042617.14 |
Clcnka
|
chloride channel, voltage-sensitive Ka |
| chr17_-_37178079 | 0.03 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr3_-_89294430 | 0.03 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr1_+_171594690 | 0.03 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr3_-_151899470 | 0.03 |
ENSMUST00000050073.13
|
Dnajb4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr5_+_124065481 | 0.03 |
ENSMUST00000166129.5
|
Gm43518
|
predicted gene 43518 |
| chr11_-_120238917 | 0.03 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr9_+_21336300 | 0.02 |
ENSMUST00000172482.8
ENSMUST00000174050.8 |
Dnm2
|
dynamin 2 |
| chr11_-_95966407 | 0.02 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr10_-_107330580 | 0.02 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chr9_-_119548388 | 0.02 |
ENSMUST00000213392.2
|
Scn10a
|
sodium channel, voltage-gated, type X, alpha |
| chr8_-_3767547 | 0.02 |
ENSMUST00000058040.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr14_+_63235512 | 0.02 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chr6_-_35303057 | 0.02 |
ENSMUST00000051176.9
|
Fam180a
|
family with sequence similarity 180, member A |
| chr1_-_139304832 | 0.02 |
ENSMUST00000198445.5
|
Crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr10_-_53252210 | 0.02 |
ENSMUST00000095691.7
|
Cep85l
|
centrosomal protein 85-like |
| chr2_+_148592272 | 0.02 |
ENSMUST00000109955.10
ENSMUST00000109954.8 ENSMUST00000109952.2 |
Cstl1
|
cystatin-like 1 |
| chrX_+_100427331 | 0.02 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr6_+_41139948 | 0.02 |
ENSMUST00000103275.4
|
Trbv17
|
T cell receptor beta, variable 17 |
| chr9_-_119548098 | 0.02 |
ENSMUST00000084787.6
|
Scn10a
|
sodium channel, voltage-gated, type X, alpha |
| chr6_-_138056914 | 0.02 |
ENSMUST00000171804.4
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr14_+_53279810 | 0.02 |
ENSMUST00000198439.5
ENSMUST00000103605.3 |
Trav8d-2
|
T cell receptor alpha variable 8D-2 |
| chr5_-_103777145 | 0.02 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr18_+_20798337 | 0.02 |
ENSMUST00000075312.5
|
Ttr
|
transthyretin |
| chr13_+_76115824 | 0.02 |
ENSMUST00000022081.3
|
Spata9
|
spermatogenesis associated 9 |
| chr14_+_32555912 | 0.01 |
ENSMUST00000104926.3
|
Fam170b
|
family with sequence similarity 170, member B |
| chr18_+_23548534 | 0.01 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr6_+_32027216 | 0.01 |
ENSMUST00000031778.6
|
1700012A03Rik
|
RIKEN cDNA 1700012A03 gene |
| chr7_-_74958121 | 0.01 |
ENSMUST00000085164.7
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr11_+_73819052 | 0.01 |
ENSMUST00000205791.2
|
Olfr396-ps1
|
olfactory receptor 396, pseudogene 1 |
| chr16_-_92118313 | 0.01 |
ENSMUST00000062638.8
|
Fam243
|
family with sequence similarity 243 |
| chr11_-_73408023 | 0.01 |
ENSMUST00000092921.3
|
Olfr382
|
olfactory receptor 382 |
| chr2_+_74552322 | 0.01 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4 |
| chr1_+_135727228 | 0.01 |
ENSMUST00000154463.8
ENSMUST00000139986.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr9_-_35755446 | 0.01 |
ENSMUST00000054175.7
|
Pate5
|
prostate and testis expressed 5 |
| chr4_+_99952988 | 0.01 |
ENSMUST00000039630.6
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr15_+_102862862 | 0.01 |
ENSMUST00000001701.4
|
Hoxc11
|
homeobox C11 |
| chr6_+_125298168 | 0.01 |
ENSMUST00000176365.2
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr7_+_44866095 | 0.01 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr14_+_55887631 | 0.01 |
ENSMUST00000007735.9
ENSMUST00000164809.9 ENSMUST00000228395.2 ENSMUST00000226591.2 |
Tssk4
|
testis-specific serine kinase 4 |
| chr13_-_55979191 | 0.01 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr9_+_49014020 | 0.01 |
ENSMUST00000070390.12
ENSMUST00000167095.8 |
Tmprss5
|
transmembrane protease, serine 5 (spinesin) |
| chr16_+_90627719 | 0.01 |
ENSMUST00000037539.15
ENSMUST00000231280.2 |
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr11_-_87780720 | 0.01 |
ENSMUST00000049743.2
|
Olfr462
|
olfactory receptor 462 |
| chr10_-_108846816 | 0.01 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr19_+_38121214 | 0.01 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr17_-_20334488 | 0.01 |
ENSMUST00000095637.2
|
Fpr-rs7
|
formyl peptide receptor, related sequence 7 |
| chr2_-_77000878 | 0.01 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr16_+_48637219 | 0.01 |
ENSMUST00000023328.8
|
Retnlb
|
resistin like beta |
| chr16_-_5040125 | 0.01 |
ENSMUST00000050160.6
|
AU021092
|
expressed sequence AU021092 |
| chr19_+_38121248 | 0.01 |
ENSMUST00000025956.13
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr7_+_104705455 | 0.01 |
ENSMUST00000098161.2
|
Olfr677
|
olfactory receptor 677 |
| chr16_-_50411484 | 0.01 |
ENSMUST00000062439.6
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr19_+_13610308 | 0.01 |
ENSMUST00000053113.5
|
Olfr1489
|
olfactory receptor 1489 |
| chr1_+_135727571 | 0.01 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr10_-_105410280 | 0.01 |
ENSMUST00000061506.9
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr17_-_89508103 | 0.01 |
ENSMUST00000035701.6
|
Fshr
|
follicle stimulating hormone receptor |
| chr2_+_91086489 | 0.01 |
ENSMUST00000154959.8
ENSMUST00000059566.11 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr9_-_117080869 | 0.01 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr13_-_98773862 | 0.01 |
ENSMUST00000050389.5
|
Tmem174
|
transmembrane protein 174 |
| chr19_+_22425534 | 0.01 |
ENSMUST00000235522.2
ENSMUST00000236372.2 ENSMUST00000238066.2 ENSMUST00000235780.2 ENSMUST00000236804.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr10_+_69370038 | 0.01 |
ENSMUST00000182439.8
ENSMUST00000092434.12 ENSMUST00000047061.13 ENSMUST00000092432.12 ENSMUST00000092431.12 ENSMUST00000054167.15 |
Ank3
|
ankyrin 3, epithelial |
| chr11_+_69047815 | 0.01 |
ENSMUST00000036424.3
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type |
| chr1_-_65112680 | 0.01 |
ENSMUST00000114064.3
|
Crygc
|
crystallin, gamma C |
| chr11_-_115967873 | 0.01 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr2_+_91086299 | 0.01 |
ENSMUST00000134699.8
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_-_58346806 | 0.01 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr3_-_137773149 | 0.01 |
ENSMUST00000053318.4
|
Gm5105
|
predicted gene 5105 |
| chr7_-_127020954 | 0.01 |
ENSMUST00000126756.2
|
Zfp688
|
zinc finger protein 688 |
| chr1_+_135727140 | 0.01 |
ENSMUST00000152208.8
ENSMUST00000152075.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr14_+_123897383 | 0.01 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chr2_+_65451100 | 0.01 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr19_+_55882942 | 0.01 |
ENSMUST00000142291.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr1_-_126758520 | 0.01 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr5_+_139338362 | 0.01 |
ENSMUST00000031521.13
|
Cyp2w1
|
cytochrome P450, family 2, subfamily w, polypeptide 1 |
| chr2_-_111814442 | 0.00 |
ENSMUST00000207885.3
|
Olfr1309
|
olfactory receptor 1309 |
| chr9_-_39149727 | 0.00 |
ENSMUST00000073248.2
|
Olfr1537
|
olfactory receptor 1537 |
| chr2_+_25179903 | 0.00 |
ENSMUST00000028337.7
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr10_+_69369632 | 0.00 |
ENSMUST00000182155.8
ENSMUST00000183169.8 ENSMUST00000183148.8 |
Ank3
|
ankyrin 3, epithelial |
| chr12_-_118930130 | 0.00 |
ENSMUST00000035515.5
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr8_+_72811504 | 0.00 |
ENSMUST00000093434.5
|
Olfr372
|
olfactory receptor 372 |
| chr7_+_103595775 | 0.00 |
ENSMUST00000098188.2
|
Olfr633
|
olfactory receptor 633 |
| chr16_+_90628001 | 0.00 |
ENSMUST00000099543.10
|
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr9_+_37903788 | 0.00 |
ENSMUST00000074611.3
|
Olfr881
|
olfactory receptor 881 |
| chr9_+_38435026 | 0.00 |
ENSMUST00000074987.3
|
Olfr911-ps1
|
olfactory receptor 911, pseudogene 1 |
| chr18_+_37822865 | 0.00 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2 |
| chr2_-_88175071 | 0.00 |
ENSMUST00000137895.3
|
Olfr1177-ps
|
olfactory receptor 1177, pseudogene |
| chr16_+_17716480 | 0.00 |
ENSMUST00000055374.8
|
Tssk2
|
testis-specific serine kinase 2 |
| chr1_+_135764092 | 0.00 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr2_-_89170692 | 0.00 |
ENSMUST00000099784.5
|
Olfr1233
|
olfactory receptor 1233 |
| chr13_-_114595122 | 0.00 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr7_+_102526329 | 0.00 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chr7_-_30623592 | 0.00 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr2_-_103133524 | 0.00 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr17_+_33418036 | 0.00 |
ENSMUST00000112165.4
|
Olfr239
|
olfactory receptor 239 |
| chr14_+_8293646 | 0.00 |
ENSMUST00000035250.6
|
Olfr720
|
olfactory receptor 720 |
| chr9_+_39305089 | 0.00 |
ENSMUST00000078531.2
|
Olfr951
|
olfactory receptor 951 |
| chr2_-_65397809 | 0.00 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr1_-_75110511 | 0.00 |
ENSMUST00000027405.6
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr6_+_40548291 | 0.00 |
ENSMUST00000051540.5
|
Olfr460
|
olfactory receptor 460 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pgr_Nr3c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.4 | 1.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.3 | 1.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.6 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.9 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.6 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.6 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.1 | 0.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.7 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0072313 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.3 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.3 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.3 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.6 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.0 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 0.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 1.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.2 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |