Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Phox2a
Z-value: 0.91
Transcription factors associated with Phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Phox2a
|
ENSMUSG00000007946.12 | paired-like homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Phox2a | mm39_v1_chr7_+_101467512_101467529 | 0.58 | 3.0e-01 | Click! |
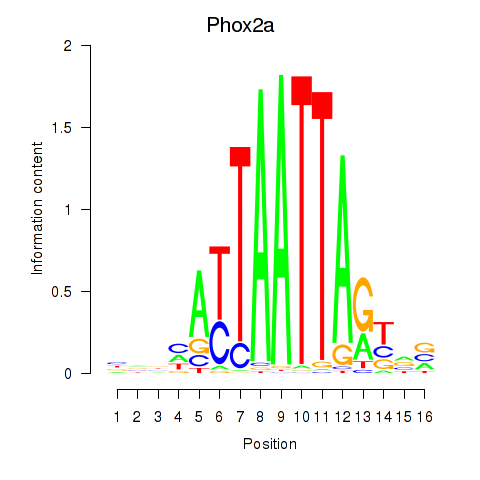
Activity profile of Phox2a motif
Sorted Z-values of Phox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89855921 | 1.70 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr9_+_118892497 | 1.34 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chrX_+_16485937 | 0.85 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr2_-_111843053 | 0.84 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chrM_+_2743 | 0.81 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr2_-_111820618 | 0.78 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr1_+_173983199 | 0.65 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr2_-_34990689 | 0.47 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr15_-_66985760 | 0.46 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr19_+_12364643 | 0.41 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr7_-_12829100 | 0.25 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr4_+_134658209 | 0.23 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr11_-_99134885 | 0.23 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chrX_+_159551009 | 0.22 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr7_-_44753168 | 0.19 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr15_+_98350469 | 0.17 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr4_+_108576846 | 0.11 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr15_+_39522905 | 0.09 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_101486983 | 0.08 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr11_+_116734104 | 0.07 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr5_+_13448833 | 0.05 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr10_-_128885867 | 0.04 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr1_+_173924457 | 0.03 |
ENSMUST00000213832.2
|
Olfr427
|
olfactory receptor 427 |
| chr6_+_37847721 | 0.03 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr5_-_118382926 | 0.02 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr10_-_129965752 | 0.02 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr12_+_30934320 | 0.01 |
ENSMUST00000067087.7
|
Alkal2
|
ALK and LTK ligand 2 |
| chr8_-_3675024 | 0.01 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr1_-_154692678 | 0.01 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr6_-_41752111 | 0.01 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr19_-_47680528 | 0.00 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr2_-_164427367 | 0.00 |
ENSMUST00000109342.2
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Phox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.8 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 1.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.0 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 4.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |