Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
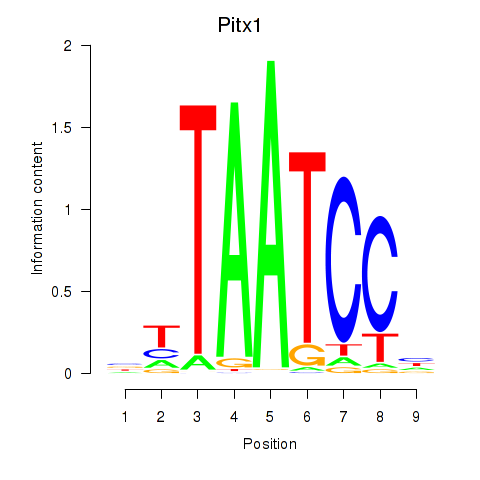
Results for Pitx1
Z-value: 0.67
Transcription factors associated with Pitx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx1
|
ENSMUSG00000021506.8 | paired-like homeodomain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx1 | mm39_v1_chr13_-_55979191_55979238 | -0.78 | 1.2e-01 | Click! |
Activity profile of Pitx1 motif
Sorted Z-values of Pitx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_109540201 | 0.64 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr4_-_156340276 | 0.53 |
ENSMUST00000220228.2
ENSMUST00000218788.2 ENSMUST00000179919.3 |
Samd11
|
sterile alpha motif domain containing 11 |
| chr10_+_80765900 | 0.48 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chrM_+_8603 | 0.45 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr7_+_16515265 | 0.40 |
ENSMUST00000108496.9
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr1_+_177272297 | 0.36 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr17_+_34124078 | 0.35 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr14_+_26722319 | 0.35 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chrX_+_168662592 | 0.35 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr11_-_68864666 | 0.32 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chrX_+_133657312 | 0.32 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr1_+_177272215 | 0.29 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_-_96615138 | 0.29 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr9_+_109704609 | 0.29 |
ENSMUST00000094324.8
|
Cdc25a
|
cell division cycle 25A |
| chr3_+_41517790 | 0.28 |
ENSMUST00000163764.8
|
Jade1
|
jade family PHD finger 1 |
| chr11_+_23206001 | 0.28 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr14_-_31362835 | 0.28 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr5_-_134485430 | 0.27 |
ENSMUST00000200944.4
ENSMUST00000202280.4 ENSMUST00000074114.12 ENSMUST00000100654.10 ENSMUST00000100650.10 ENSMUST00000111245.9 ENSMUST00000202554.4 ENSMUST00000073161.12 ENSMUST00000100652.10 ENSMUST00000171794.9 ENSMUST00000167084.9 |
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr3_+_41517829 | 0.27 |
ENSMUST00000170711.8
|
Jade1
|
jade family PHD finger 1 |
| chrX_+_168468186 | 0.27 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr3_+_106020545 | 0.26 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr11_+_87938626 | 0.26 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr19_-_38032006 | 0.25 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr10_-_115198093 | 0.25 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr2_-_23939401 | 0.25 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr11_+_3280771 | 0.24 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_-_126342551 | 0.23 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr19_-_4976844 | 0.23 |
ENSMUST00000236496.2
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr11_+_29668563 | 0.23 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr5_+_75735576 | 0.22 |
ENSMUST00000144270.8
ENSMUST00000005815.7 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr11_+_90078485 | 0.22 |
ENSMUST00000124877.3
ENSMUST00000153734.2 ENSMUST00000134929.8 |
Gm45716
|
predicted gene 45716 |
| chr19_-_47907705 | 0.22 |
ENSMUST00000095998.7
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr18_+_36812246 | 0.21 |
ENSMUST00000186538.7
|
Slc35a4
|
solute carrier family 35, member A4 |
| chr11_-_22932090 | 0.21 |
ENSMUST00000160826.2
ENSMUST00000093270.6 ENSMUST00000071068.9 ENSMUST00000159081.8 |
Commd1b
Commd1
|
COMM domain containing 1B COMM domain containing 1 |
| chr7_-_109092834 | 0.20 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr11_-_59078565 | 0.19 |
ENSMUST00000170895.3
|
Guk1
|
guanylate kinase 1 |
| chr15_+_80832685 | 0.19 |
ENSMUST00000023043.10
ENSMUST00000164806.6 ENSMUST00000207170.2 ENSMUST00000168756.8 |
Adsl
|
adenylosuccinate lyase |
| chrX_+_73468140 | 0.19 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr5_-_23821523 | 0.19 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr19_-_38031774 | 0.19 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chrX_-_135642025 | 0.18 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr19_+_9957940 | 0.18 |
ENSMUST00000235196.2
|
Fth1
|
ferritin heavy polypeptide 1 |
| chr6_+_58810674 | 0.18 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr4_+_102446883 | 0.18 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr11_+_87938128 | 0.18 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr11_+_87938519 | 0.17 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr1_-_171050004 | 0.17 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr5_+_86219593 | 0.17 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr3_+_102965910 | 0.17 |
ENSMUST00000199367.5
ENSMUST00000199049.5 ENSMUST00000197678.5 |
Nras
|
neuroblastoma ras oncogene |
| chr8_+_79236051 | 0.17 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr10_-_115197775 | 0.17 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr16_-_30900181 | 0.16 |
ENSMUST00000055389.9
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr7_-_25098122 | 0.16 |
ENSMUST00000105177.3
ENSMUST00000149349.2 |
Lipe
|
lipase, hormone sensitive |
| chr8_+_85786684 | 0.16 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr1_-_80255156 | 0.16 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr9_-_108443916 | 0.16 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chrX_-_135877607 | 0.16 |
ENSMUST00000127404.8
ENSMUST00000113071.2 |
Tmsb15l
Tmsb15b1
|
thymosin beta 15b like thymosin beta 15b1 |
| chrX_+_20291927 | 0.15 |
ENSMUST00000115384.9
|
Jade3
|
jade family PHD finger 3 |
| chr11_+_95275458 | 0.14 |
ENSMUST00000021243.16
ENSMUST00000146556.2 |
Slc35b1
|
solute carrier family 35, member B1 |
| chrX_-_135641869 | 0.14 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr5_-_125371162 | 0.14 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr14_+_52348396 | 0.14 |
ENSMUST00000111600.11
|
Rpgrip1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr1_-_171050077 | 0.14 |
ENSMUST00000005817.9
|
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr7_-_4517367 | 0.13 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr11_+_19874403 | 0.13 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr3_+_136375839 | 0.12 |
ENSMUST00000070198.14
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr2_+_120307390 | 0.12 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr10_-_9776823 | 0.12 |
ENSMUST00000141722.8
|
Stxbp5
|
syntaxin binding protein 5 (tomosyn) |
| chr3_+_136376440 | 0.11 |
ENSMUST00000056758.9
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr18_+_46730765 | 0.11 |
ENSMUST00000238168.2
ENSMUST00000078079.11 ENSMUST00000168382.2 ENSMUST00000235849.2 ENSMUST00000235973.2 ENSMUST00000235455.2 ENSMUST00000237478.2 |
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr9_-_100368841 | 0.11 |
ENSMUST00000098458.4
|
Il20rb
|
interleukin 20 receptor beta |
| chr9_-_109930078 | 0.11 |
ENSMUST00000196455.2
ENSMUST00000062368.13 |
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
| chr9_-_108444561 | 0.11 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr4_-_156339769 | 0.11 |
ENSMUST00000217934.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr17_+_35188888 | 0.11 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr11_-_107228382 | 0.11 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr17_+_28259749 | 0.10 |
ENSMUST00000233869.2
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr2_-_121287174 | 0.10 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chr9_+_75213570 | 0.10 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr1_-_165762469 | 0.10 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr3_+_88523730 | 0.10 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr4_-_42661893 | 0.10 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr5_+_3393893 | 0.09 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chrX_-_135858748 | 0.09 |
ENSMUST00000069803.5
|
Tmsb15b2
|
thymosin beta 15b2 |
| chr19_-_32689687 | 0.09 |
ENSMUST00000237752.2
ENSMUST00000235412.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr3_+_105881027 | 0.09 |
ENSMUST00000000573.9
|
Ovgp1
|
oviductal glycoprotein 1 |
| chr5_+_130215996 | 0.08 |
ENSMUST00000119027.8
ENSMUST00000026390.8 |
Rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr11_+_23206565 | 0.08 |
ENSMUST00000136235.2
|
Xpo1
|
exportin 1 |
| chr12_+_8971603 | 0.08 |
ENSMUST00000020909.4
|
Laptm4a
|
lysosomal-associated protein transmembrane 4A |
| chr3_+_94217396 | 0.08 |
ENSMUST00000049822.10
|
Them4
|
thioesterase superfamily member 4 |
| chr4_+_116565898 | 0.08 |
ENSMUST00000135499.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr19_+_36061096 | 0.08 |
ENSMUST00000025714.9
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr11_+_19874354 | 0.08 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr11_+_116547932 | 0.07 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr4_-_109333866 | 0.07 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr12_+_79255678 | 0.07 |
ENSMUST00000021548.12
|
Rdh12
|
retinol dehydrogenase 12 |
| chr14_-_64654397 | 0.07 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr19_-_12742811 | 0.07 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr14_-_121976273 | 0.07 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr5_-_92583078 | 0.07 |
ENSMUST00000038514.15
|
Nup54
|
nucleoporin 54 |
| chr5_-_137785903 | 0.07 |
ENSMUST00000196022.2
|
Mepce
|
methylphosphate capping enzyme |
| chr10_+_21869776 | 0.07 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_+_18283405 | 0.07 |
ENSMUST00000037341.14
|
Nhsl1
|
NHS-like 1 |
| chr2_+_101455079 | 0.07 |
ENSMUST00000111227.2
|
Rag2
|
recombination activating gene 2 |
| chr7_+_24206431 | 0.06 |
ENSMUST00000176880.2
|
Zfp428
|
zinc finger protein 428 |
| chr10_-_6930376 | 0.06 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr11_+_69886652 | 0.06 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr7_+_5065142 | 0.06 |
ENSMUST00000005041.15
ENSMUST00000209099.2 |
U2af2
|
U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 2 |
| chr5_-_134485081 | 0.06 |
ENSMUST00000111244.5
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr8_+_79235946 | 0.06 |
ENSMUST00000209490.2
ENSMUST00000034111.10 |
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr14_-_20027112 | 0.06 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr5_-_144160397 | 0.06 |
ENSMUST00000085701.7
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chrX_-_42256694 | 0.06 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr2_-_145776934 | 0.06 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr7_-_4517608 | 0.06 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr17_+_34092811 | 0.06 |
ENSMUST00000226885.2
|
Smim40-ps
|
small integral membrane protein 40, pseudogene |
| chrX_+_92718695 | 0.05 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_-_97242842 | 0.05 |
ENSMUST00000093942.5
|
Gpr179
|
G protein-coupled receptor 179 |
| chr10_-_125164399 | 0.05 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr2_+_69500444 | 0.05 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr13_+_94925382 | 0.05 |
ENSMUST00000046644.7
|
Tbca
|
tubulin cofactor A |
| chr19_+_8861096 | 0.05 |
ENSMUST00000187504.7
|
Lbhd1
|
LBH domain containing 1 |
| chr17_+_57077258 | 0.05 |
ENSMUST00000168666.3
|
Prr22
|
proline rich 22 |
| chr9_+_35334878 | 0.05 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr1_-_4430481 | 0.05 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chrX_+_55825033 | 0.04 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr12_+_79255702 | 0.04 |
ENSMUST00000122227.8
|
Rdh12
|
retinol dehydrogenase 12 |
| chr11_-_99383938 | 0.04 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr8_+_12965876 | 0.04 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr19_-_6127211 | 0.04 |
ENSMUST00000160590.2
ENSMUST00000025711.13 |
Vps51
|
VPS51 GARP complex subunit |
| chr2_+_89642395 | 0.04 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr9_+_50528608 | 0.04 |
ENSMUST00000000171.15
ENSMUST00000151197.8 |
Pih1d2
|
PIH1 domain containing 2 |
| chr8_-_106863521 | 0.04 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr3_-_9029097 | 0.04 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr2_+_124910037 | 0.04 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr10_+_25284244 | 0.04 |
ENSMUST00000092645.7
ENSMUST00000219805.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr14_+_59438879 | 0.04 |
ENSMUST00000140136.9
ENSMUST00000142326.2 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr11_+_97917520 | 0.04 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19 |
| chr3_+_88523440 | 0.04 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr5_-_90487583 | 0.04 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr7_-_4517559 | 0.04 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr11_-_29197222 | 0.04 |
ENSMUST00000020754.10
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr2_-_101459274 | 0.04 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chrX_-_99670174 | 0.04 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chr9_+_35332837 | 0.04 |
ENSMUST00000119129.10
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr1_+_9615619 | 0.04 |
ENSMUST00000072079.9
|
Rrs1
|
ribosome biogenesis regulator 1 |
| chr12_-_114579763 | 0.03 |
ENSMUST00000103500.2
|
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr12_-_102406069 | 0.03 |
ENSMUST00000021607.9
|
Lgmn
|
legumain |
| chr1_+_12762501 | 0.03 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chrX_+_133486391 | 0.03 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr7_-_132616977 | 0.03 |
ENSMUST00000169570.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr9_+_108367801 | 0.03 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr15_-_101801351 | 0.03 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chrX_+_7473338 | 0.03 |
ENSMUST00000115726.9
ENSMUST00000133637.8 ENSMUST00000115725.9 |
Cacna1f
|
calcium channel, voltage-dependent, alpha 1F subunit |
| chr1_-_84818223 | 0.03 |
ENSMUST00000186465.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr10_-_80257681 | 0.03 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chr11_+_4587733 | 0.03 |
ENSMUST00000070257.14
ENSMUST00000109930.3 |
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr6_-_93890520 | 0.03 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr17_-_59298338 | 0.02 |
ENSMUST00000025064.14
|
Pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr12_-_118265103 | 0.02 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr16_+_32066037 | 0.02 |
ENSMUST00000141820.8
ENSMUST00000178573.8 ENSMUST00000023474.4 ENSMUST00000135289.2 |
Wdr53
|
WD repeat domain 53 |
| chr15_+_91722524 | 0.02 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr2_+_22958179 | 0.02 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr4_+_19280850 | 0.02 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chrX_+_162923474 | 0.02 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr4_+_80752360 | 0.02 |
ENSMUST00000133655.8
ENSMUST00000006151.13 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr17_-_57307018 | 0.02 |
ENSMUST00000002740.3
|
Pspn
|
persephin |
| chr9_-_71070506 | 0.02 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr4_-_140780972 | 0.02 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chrX_+_55824797 | 0.02 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_-_87240405 | 0.02 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr2_+_119156265 | 0.02 |
ENSMUST00000102517.4
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr10_-_129023288 | 0.02 |
ENSMUST00000072976.4
|
Olfr773
|
olfactory receptor 773 |
| chr5_-_86780277 | 0.02 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr10_-_18890281 | 0.02 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_+_12762712 | 0.02 |
ENSMUST00000186051.7
|
Sulf1
|
sulfatase 1 |
| chr19_-_32173824 | 0.02 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr4_+_80752535 | 0.02 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr3_-_144555062 | 0.01 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr7_+_126575510 | 0.01 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr5_+_88523967 | 0.01 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr7_+_88079534 | 0.01 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chr2_+_136734088 | 0.01 |
ENSMUST00000099311.9
|
Slx4ip
|
SLX4 interacting protein |
| chr10_+_90412827 | 0.01 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_+_67167950 | 0.01 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr14_-_8457069 | 0.01 |
ENSMUST00000022257.4
|
Atxn7
|
ataxin 7 |
| chr14_-_49482846 | 0.01 |
ENSMUST00000227113.2
ENSMUST00000130853.2 ENSMUST00000228936.2 ENSMUST00000022398.15 |
ccdc198
|
coiled-coil domain containing 198 |
| chr9_+_32135540 | 0.01 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_-_113047397 | 0.01 |
ENSMUST00000080673.13
ENSMUST00000208151.2 ENSMUST00000208290.2 |
Ryr3
|
ryanodine receptor 3 |
| chr1_+_44022321 | 0.01 |
ENSMUST00000190207.2
|
Tpp2
|
tripeptidyl peptidase II |
| chr3_+_121838076 | 0.01 |
ENSMUST00000013995.13
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr2_+_85615818 | 0.01 |
ENSMUST00000047870.6
|
Olfr1015
|
olfactory receptor 1015 |
| chr11_+_69886603 | 0.01 |
ENSMUST00000018716.10
|
Phf23
|
PHD finger protein 23 |
| chr11_+_94901104 | 0.01 |
ENSMUST00000124735.2
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr4_+_101407608 | 0.01 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr13_-_105431153 | 0.01 |
ENSMUST00000224749.2
|
Rnf180
|
ring finger protein 180 |
| chr15_+_91722458 | 0.01 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr16_-_26786995 | 0.01 |
ENSMUST00000231969.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr13_-_105430932 | 0.01 |
ENSMUST00000224662.2
|
Rnf180
|
ring finger protein 180 |
| chr9_+_38240356 | 0.01 |
ENSMUST00000214155.2
ENSMUST00000212354.3 |
Olfr25
|
olfactory receptor 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.3 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.2 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.2 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.2 | GO:1904349 | positive regulation of small intestine smooth muscle contraction(GO:1904349) |
| 0.0 | 0.7 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 0.1 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |