Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
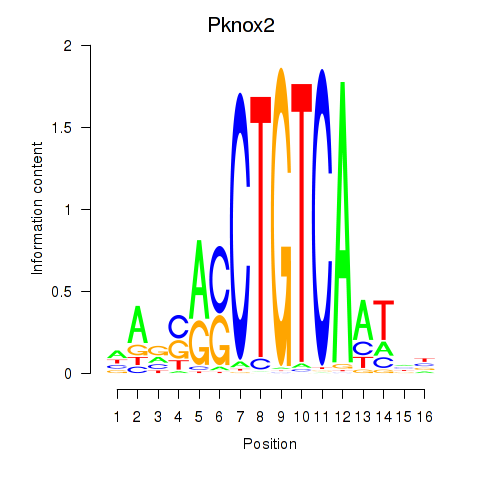
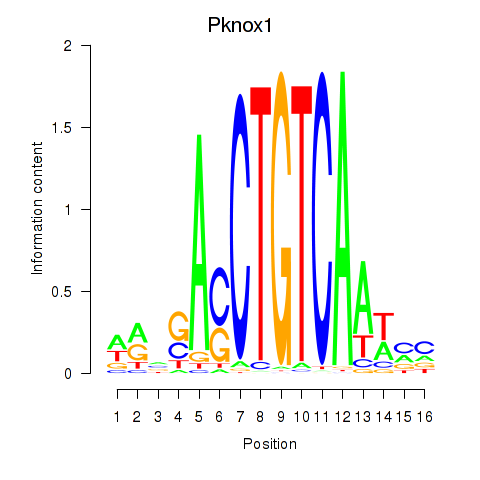
Results for Pknox2_Pknox1
Z-value: 0.28
Transcription factors associated with Pknox2_Pknox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pknox2
|
ENSMUSG00000035934.17 | Pbx/knotted 1 homeobox 2 |
|
Pknox1
|
ENSMUSG00000006705.14 | Pbx/knotted 1 homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pknox1 | mm39_v1_chr17_+_31810693_31810729 | 0.95 | 1.4e-02 | Click! |
| Pknox2 | mm39_v1_chr9_-_37058590_37058703 | -0.49 | 4.0e-01 | Click! |
Activity profile of Pknox2_Pknox1 motif
Sorted Z-values of Pknox2_Pknox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_23206001 | 0.22 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr7_-_126873219 | 0.19 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr2_-_34803988 | 0.16 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr11_+_117877118 | 0.13 |
ENSMUST00000132676.8
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr1_+_156386414 | 0.12 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr18_+_56565188 | 0.12 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr16_-_95928804 | 0.12 |
ENSMUST00000233292.2
ENSMUST00000050884.16 |
Hmgn1
|
high mobility group nucleosomal binding domain 1 |
| chr3_+_121243395 | 0.10 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr16_+_22926162 | 0.10 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_+_98352298 | 0.10 |
ENSMUST00000033009.16
|
Thap12
|
THAP domain containing 12 |
| chr17_+_34423054 | 0.09 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr3_+_95139598 | 0.09 |
ENSMUST00000107200.8
ENSMUST00000107199.2 |
Cdc42se1
|
CDC42 small effector 1 |
| chr17_-_71158184 | 0.09 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr12_-_85420684 | 0.09 |
ENSMUST00000222585.2
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr3_+_51324022 | 0.09 |
ENSMUST00000192419.6
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr11_+_78215026 | 0.08 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr2_+_30306116 | 0.08 |
ENSMUST00000113601.10
ENSMUST00000113603.10 |
Ptpa
|
protein phosphatase 2 protein activator |
| chr8_+_89423645 | 0.08 |
ENSMUST00000043526.15
ENSMUST00000211554.2 ENSMUST00000209532.2 ENSMUST00000209559.2 |
Cyld
|
CYLD lysine 63 deubiquitinase |
| chr1_+_134110142 | 0.07 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr4_+_133970973 | 0.07 |
ENSMUST00000135228.8
ENSMUST00000144222.8 ENSMUST00000143448.8 ENSMUST00000125921.8 ENSMUST00000122952.8 ENSMUST00000131447.2 |
E130218I03Rik
|
RIKEN cDNA E130218I03 gene |
| chr12_-_112893382 | 0.07 |
ENSMUST00000075827.5
|
Jag2
|
jagged 2 |
| chr6_+_95094721 | 0.07 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr1_-_37580084 | 0.06 |
ENSMUST00000151952.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr16_-_10993919 | 0.06 |
ENSMUST00000142389.3
ENSMUST00000138185.9 |
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr12_-_79211820 | 0.06 |
ENSMUST00000162569.8
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr9_+_113760002 | 0.06 |
ENSMUST00000084885.12
ENSMUST00000009885.14 |
Ubp1
|
upstream binding protein 1 |
| chr10_+_82534841 | 0.06 |
ENSMUST00000020478.14
|
Hcfc2
|
host cell factor C2 |
| chr3_-_108352459 | 0.06 |
ENSMUST00000132467.8
ENSMUST00000102625.11 |
Sars
|
seryl-aminoacyl-tRNA synthetase |
| chr17_-_71158703 | 0.06 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr5_+_35146727 | 0.06 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr14_+_55252911 | 0.06 |
ENSMUST00000022815.10
|
Ngdn
|
neuroguidin, EIF4E binding protein |
| chr16_-_10993386 | 0.05 |
ENSMUST00000125537.9
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr7_-_45083688 | 0.05 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr15_+_88703786 | 0.05 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr5_-_109706801 | 0.05 |
ENSMUST00000200284.5
ENSMUST00000044579.12 |
Crlf2
|
cytokine receptor-like factor 2 |
| chr17_+_30843328 | 0.05 |
ENSMUST00000170651.2
|
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr9_-_71678814 | 0.05 |
ENSMUST00000122065.2
ENSMUST00000121322.8 ENSMUST00000072899.9 |
Cgnl1
|
cingulin-like 1 |
| chr9_+_68561042 | 0.05 |
ENSMUST00000034766.14
|
Rora
|
RAR-related orphan receptor alpha |
| chr1_-_36283326 | 0.05 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr19_-_9876745 | 0.05 |
ENSMUST00000237725.2
|
Incenp
|
inner centromere protein |
| chr3_+_51323383 | 0.04 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr1_+_134109888 | 0.04 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr6_-_106777014 | 0.04 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chr4_+_11758147 | 0.04 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr5_+_35146880 | 0.04 |
ENSMUST00000114285.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_+_34919078 | 0.04 |
ENSMUST00000080036.3
|
Htt
|
huntingtin |
| chr5_+_24679154 | 0.04 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr7_-_126483851 | 0.04 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr5_+_93416091 | 0.04 |
ENSMUST00000121127.2
|
Ccng2
|
cyclin G2 |
| chr1_+_74627506 | 0.04 |
ENSMUST00000113732.2
|
Bcs1l
|
BCS1-like (yeast) |
| chr5_-_110434026 | 0.04 |
ENSMUST00000031472.12
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr7_+_65693350 | 0.04 |
ENSMUST00000206065.2
|
Gm45213
|
predicted gene 45213 |
| chr3_-_51468236 | 0.04 |
ENSMUST00000037141.9
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr4_+_126915104 | 0.03 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr17_+_30845909 | 0.03 |
ENSMUST00000236140.2
ENSMUST00000236118.2 ENSMUST00000235390.2 |
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr17_-_31738901 | 0.03 |
ENSMUST00000171291.2
ENSMUST00000237807.2 |
Wdr4
|
WD repeat domain 4 |
| chr16_+_91188609 | 0.03 |
ENSMUST00000160764.2
|
Gm21970
|
predicted gene 21970 |
| chr11_-_79971750 | 0.03 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr3_+_137570248 | 0.03 |
ENSMUST00000041045.14
|
H2az1
|
H2A.Z variant histone 1 |
| chr13_+_76532470 | 0.03 |
ENSMUST00000125209.8
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr15_-_76608009 | 0.03 |
ENSMUST00000036247.10
|
C030006K11Rik
|
RIKEN cDNA C030006K11 gene |
| chr17_-_30845845 | 0.03 |
ENSMUST00000235547.2
ENSMUST00000188852.2 |
Glo1
1700097N02Rik
|
glyoxalase 1 RIKEN cDNA 1700097N02 gene |
| chr11_-_4544751 | 0.03 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr10_-_123032821 | 0.03 |
ENSMUST00000219619.2
ENSMUST00000020334.9 |
Usp15
|
ubiquitin specific peptidase 15 |
| chr17_-_25155868 | 0.03 |
ENSMUST00000115228.9
ENSMUST00000117509.8 ENSMUST00000121723.8 ENSMUST00000119115.8 ENSMUST00000121787.8 ENSMUST00000088345.12 ENSMUST00000120035.8 ENSMUST00000115229.10 ENSMUST00000178969.8 |
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr12_-_108860042 | 0.03 |
ENSMUST00000109848.10
|
Wars
|
tryptophanyl-tRNA synthetase |
| chr7_-_45084012 | 0.03 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr4_+_3938881 | 0.03 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr2_+_160487801 | 0.02 |
ENSMUST00000109468.3
|
Top1
|
topoisomerase (DNA) I |
| chr16_+_55794244 | 0.02 |
ENSMUST00000050248.9
|
Zbtb11
|
zinc finger and BTB domain containing 11 |
| chr1_+_85721132 | 0.02 |
ENSMUST00000113360.8
ENSMUST00000126962.3 |
Cab39
|
calcium binding protein 39 |
| chr3_+_85481416 | 0.02 |
ENSMUST00000107672.8
ENSMUST00000127348.8 ENSMUST00000107674.2 |
Gatb
|
glutamyl-tRNA(Gln) amidotransferase, subunit B |
| chr7_+_45084300 | 0.02 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr5_-_138617915 | 0.02 |
ENSMUST00000129832.2
|
Zfp68
|
zinc finger protein 68 |
| chr10_-_62486948 | 0.02 |
ENSMUST00000020270.6
|
Ddx50
|
DExD box helicase 50 |
| chr3_-_107538996 | 0.02 |
ENSMUST00000064759.7
|
Strip1
|
striatin interacting protein 1 |
| chr1_-_180023518 | 0.02 |
ENSMUST00000162769.8
ENSMUST00000161379.2 ENSMUST00000027766.13 ENSMUST00000161814.8 |
Coq8a
|
coenzyme Q8A |
| chr18_-_36201664 | 0.02 |
ENSMUST00000239409.2
|
Nrg2
|
neuregulin 2 |
| chr10_+_34173426 | 0.02 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr4_-_108158242 | 0.02 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr5_-_138617952 | 0.02 |
ENSMUST00000063262.11
ENSMUST00000085852.11 ENSMUST00000110905.9 |
Zfp68
|
zinc finger protein 68 |
| chr7_-_12743961 | 0.02 |
ENSMUST00000210108.2
|
Zbtb45
|
zinc finger and BTB domain containing 45 |
| chr7_-_131012051 | 0.02 |
ENSMUST00000154602.8
ENSMUST00000142349.3 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr2_+_163837423 | 0.02 |
ENSMUST00000131288.2
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr7_-_126183392 | 0.02 |
ENSMUST00000128970.8
ENSMUST00000116269.9 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chrX_-_108056995 | 0.02 |
ENSMUST00000033597.9
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr1_+_156443472 | 0.01 |
ENSMUST00000190749.2
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr11_+_32405367 | 0.01 |
ENSMUST00000051053.5
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr14_+_55813074 | 0.01 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr4_+_3938903 | 0.01 |
ENSMUST00000121210.8
ENSMUST00000121651.8 ENSMUST00000041122.11 ENSMUST00000120732.8 ENSMUST00000119307.8 ENSMUST00000123769.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr7_+_45084257 | 0.01 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr10_-_63174801 | 0.01 |
ENSMUST00000020257.13
ENSMUST00000177694.8 ENSMUST00000105442.3 |
Sirt1
|
sirtuin 1 |
| chr11_+_72687080 | 0.01 |
ENSMUST00000207107.2
|
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chrX_-_101548965 | 0.01 |
ENSMUST00000087916.11
|
Hdac8
|
histone deacetylase 8 |
| chr17_-_57024660 | 0.01 |
ENSMUST00000164907.3
ENSMUST00000233208.2 |
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr1_-_52856662 | 0.01 |
ENSMUST00000162576.8
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr11_-_97913420 | 0.01 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chrX_+_13499008 | 0.01 |
ENSMUST00000096492.4
|
Gpr34
|
G protein-coupled receptor 34 |
| chr10_-_129781929 | 0.01 |
ENSMUST00000074308.3
|
Olfr818
|
olfactory receptor 818 |
| chr10_-_123032881 | 0.01 |
ENSMUST00000220377.2
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr15_+_32920869 | 0.01 |
ENSMUST00000022871.7
|
Sdc2
|
syndecan 2 |
| chr17_-_65920481 | 0.01 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr17_+_57024759 | 0.01 |
ENSMUST00000002452.8
ENSMUST00000233832.2 ENSMUST00000233233.2 |
Ndufa11
|
NADH:ubiquinone oxidoreductase subunit A11 |
| chr2_+_30282414 | 0.01 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr17_-_32643067 | 0.01 |
ENSMUST00000237130.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr13_-_64300964 | 0.01 |
ENSMUST00000059817.12
ENSMUST00000117241.2 |
Zfp367
|
zinc finger protein 367 |
| chr14_-_70526726 | 0.01 |
ENSMUST00000228911.2
|
Ppp3cc
|
protein phosphatase 3, catalytic subunit, gamma isoform |
| chr2_+_30282266 | 0.00 |
ENSMUST00000028209.15
|
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr8_-_107007596 | 0.00 |
ENSMUST00000212896.2
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr1_+_156193607 | 0.00 |
ENSMUST00000102782.4
|
Gm2000
|
predicted gene 2000 |
| chr17_+_35844091 | 0.00 |
ENSMUST00000025273.9
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human) |
| chr10_-_7668560 | 0.00 |
ENSMUST00000065124.2
|
Ginm1
|
glycoprotein integral membrane 1 |
| chr10_-_129574784 | 0.00 |
ENSMUST00000095245.2
|
Olfr806
|
olfactory receptor 806 |
| chr4_+_41465134 | 0.00 |
ENSMUST00000030154.7
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr1_+_74627445 | 0.00 |
ENSMUST00000113733.10
ENSMUST00000027358.11 |
Bcs1l
|
BCS1-like (yeast) |
| chr10_-_117546660 | 0.00 |
ENSMUST00000020408.16
ENSMUST00000105263.9 |
Mdm2
|
transformed mouse 3T3 cell double minute 2 |
| chrX_-_47123719 | 0.00 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr11_-_20781009 | 0.00 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr19_-_44058175 | 0.00 |
ENSMUST00000172041.8
ENSMUST00000071698.13 ENSMUST00000112028.10 |
Erlin1
|
ER lipid raft associated 1 |
| chr1_+_106866678 | 0.00 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr7_+_24181416 | 0.00 |
ENSMUST00000068023.8
|
Cadm4
|
cell adhesion molecule 4 |
| chr7_-_141794815 | 0.00 |
ENSMUST00000211591.2
|
Gm40460
|
predicted gene, 40460 |
| chr2_-_36703102 | 0.00 |
ENSMUST00000100151.2
|
Olfr3
|
olfactory receptor 3 |
| chr2_+_111610658 | 0.00 |
ENSMUST00000054004.2
|
Olfr1302
|
olfactory receptor 1302 |
| chr7_+_104529916 | 0.00 |
ENSMUST00000098165.2
|
Olfr665
|
olfactory receptor 665 |
| chr2_+_162829250 | 0.00 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr14_-_52704547 | 0.00 |
ENSMUST00000205811.2
|
Olfr1508
|
olfactory receptor 1508 |
| chr19_-_6910922 | 0.00 |
ENSMUST00000235248.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr7_+_13132075 | 0.00 |
ENSMUST00000117400.2
|
Cabp5
|
calcium binding protein 5 |
| chrX_+_48552803 | 0.00 |
ENSMUST00000130558.8
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chrX_-_37131983 | 0.00 |
ENSMUST00000089060.5
|
Btg1c
|
BTG anti-proliferation factor 1C |
| chr19_-_44057800 | 0.00 |
ENSMUST00000170801.8
|
Erlin1
|
ER lipid raft associated 1 |
| chrX_-_152795636 | 0.00 |
ENSMUST00000178342.8
|
Samt1
|
spermatogenesis associated multipass transmembrane protein 1 |
| chr16_-_19079594 | 0.00 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr5_+_57879201 | 0.00 |
ENSMUST00000199310.2
|
Pcdh7
|
protocadherin 7 |
| chr1_+_135980488 | 0.00 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pknox2_Pknox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0002477 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |