Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
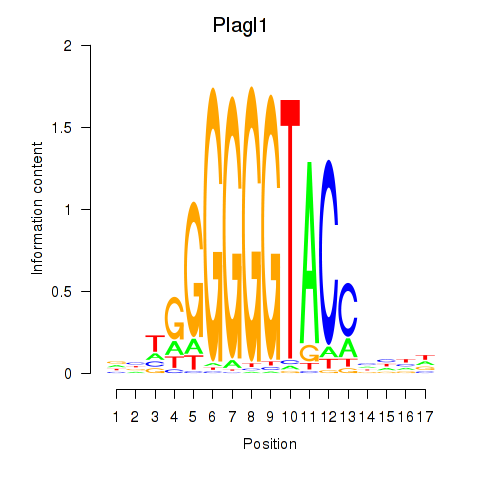
Results for Plagl1
Z-value: 1.60
Transcription factors associated with Plagl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plagl1
|
ENSMUSG00000019817.19 | pleiomorphic adenoma gene-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plagl1 | mm39_v1_chr10_+_12966532_12966587 | -0.80 | 1.0e-01 | Click! |
Activity profile of Plagl1 motif
Sorted Z-values of Plagl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_24598633 | 1.20 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr6_-_56681657 | 1.07 |
ENSMUST00000176595.3
ENSMUST00000170382.5 ENSMUST00000203958.2 |
Lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr8_-_58106027 | 0.87 |
ENSMUST00000110316.3
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_-_131987008 | 0.87 |
ENSMUST00000028815.15
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr18_+_48179711 | 0.81 |
ENSMUST00000235307.2
|
Eno1b
|
enolase 1B, retrotransposed |
| chrX_+_73348832 | 0.77 |
ENSMUST00000153141.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr17_-_26087696 | 0.75 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr10_+_61431271 | 0.74 |
ENSMUST00000020287.8
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr6_+_83156550 | 0.72 |
ENSMUST00000113919.10
ENSMUST00000113918.8 ENSMUST00000141680.8 |
Dctn1
|
dynactin 1 |
| chr14_+_71011744 | 0.72 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr1_-_131025562 | 0.70 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr8_-_58106057 | 0.68 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr6_-_128415640 | 0.68 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
| chr19_-_46327024 | 0.66 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr17_-_74602469 | 0.66 |
ENSMUST00000233144.2
|
Memo1
|
mediator of cell motility 1 |
| chrX_+_35592006 | 0.65 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr11_-_76289888 | 0.65 |
ENSMUST00000021204.4
|
Nxn
|
nucleoredoxin |
| chr15_+_102379503 | 0.62 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr6_-_128414616 | 0.62 |
ENSMUST00000151796.3
|
Fkbp4
|
FK506 binding protein 4 |
| chr6_-_134864731 | 0.61 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr5_-_124490296 | 0.60 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr17_+_29251602 | 0.60 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr1_+_134383247 | 0.58 |
ENSMUST00000112232.8
ENSMUST00000027725.11 ENSMUST00000116528.2 |
Klhl12
|
kelch-like 12 |
| chr19_-_46327071 | 0.56 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr15_+_76227695 | 0.56 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr7_+_49428082 | 0.54 |
ENSMUST00000032715.13
|
Prmt3
|
protein arginine N-methyltransferase 3 |
| chr11_+_98632953 | 0.53 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr6_-_52211882 | 0.52 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr7_-_23998735 | 0.52 |
ENSMUST00000145131.8
|
Zfp61
|
zinc finger protein 61 |
| chr12_-_13299136 | 0.51 |
ENSMUST00000221623.2
|
Ddx1
|
DEAD box helicase 1 |
| chr18_-_61533434 | 0.51 |
ENSMUST00000063307.6
ENSMUST00000075299.13 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr11_+_98632696 | 0.50 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr6_-_124689094 | 0.49 |
ENSMUST00000004379.8
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr7_-_19005721 | 0.47 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr2_-_155434487 | 0.47 |
ENSMUST00000155347.2
ENSMUST00000130881.8 ENSMUST00000079691.13 |
Gss
|
glutathione synthetase |
| chr12_-_103409912 | 0.46 |
ENSMUST00000055071.9
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr9_+_46184362 | 0.45 |
ENSMUST00000156440.8
ENSMUST00000114552.4 |
Zpr1
|
ZPR1 zinc finger |
| chr4_+_102835888 | 0.44 |
ENSMUST00000223169.2
|
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr13_-_74498320 | 0.44 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr1_+_180731843 | 0.44 |
ENSMUST00000027802.9
|
Pycr2
|
pyrroline-5-carboxylate reductase family, member 2 |
| chr1_-_131207279 | 0.44 |
ENSMUST00000062108.10
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr1_-_167112784 | 0.43 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr11_+_98632631 | 0.43 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr5_-_92231314 | 0.42 |
ENSMUST00000169094.8
ENSMUST00000167918.8 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_+_74627506 | 0.39 |
ENSMUST00000113732.2
|
Bcs1l
|
BCS1-like (yeast) |
| chr3_-_89867869 | 0.39 |
ENSMUST00000069805.14
|
Atp8b2
|
ATPase, class I, type 8B, member 2 |
| chr15_+_102379621 | 0.39 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr2_-_84500951 | 0.37 |
ENSMUST00000189988.3
ENSMUST00000189636.8 ENSMUST00000102646.4 ENSMUST00000102647.11 ENSMUST00000117299.10 |
Selenoh
|
selenoprotein H |
| chr12_-_13299197 | 0.36 |
ENSMUST00000071103.10
|
Ddx1
|
DEAD box helicase 1 |
| chr5_+_125552878 | 0.36 |
ENSMUST00000031445.5
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr11_+_32405367 | 0.36 |
ENSMUST00000051053.5
|
Ubtd2
|
ubiquitin domain containing 2 |
| chrX_-_93166992 | 0.35 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chr3_+_40754448 | 0.35 |
ENSMUST00000026858.11
|
Plk4
|
polo like kinase 4 |
| chr7_-_4518958 | 0.34 |
ENSMUST00000163710.8
ENSMUST00000166268.8 ENSMUST00000071798.13 ENSMUST00000178163.8 ENSMUST00000108587.9 |
Tnnt1
|
troponin T1, skeletal, slow |
| chr6_-_124689001 | 0.34 |
ENSMUST00000203238.2
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr9_+_108539296 | 0.32 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr17_+_29879684 | 0.31 |
ENSMUST00000235014.2
ENSMUST00000130423.4 |
Cmtr1
|
cap methyltransferase 1 |
| chr18_+_32510270 | 0.31 |
ENSMUST00000234857.2
ENSMUST00000234496.2 ENSMUST00000091967.13 ENSMUST00000025239.9 |
Bin1
|
bridging integrator 1 |
| chr19_-_42768374 | 0.31 |
ENSMUST00000069298.13
ENSMUST00000160455.8 ENSMUST00000162004.8 |
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr2_+_35472841 | 0.30 |
ENSMUST00000135741.8
|
Dab2ip
|
disabled 2 interacting protein |
| chr2_-_152672535 | 0.27 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr10_-_128383508 | 0.26 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr13_+_104365880 | 0.26 |
ENSMUST00000022227.8
|
Cenpk
|
centromere protein K |
| chr1_-_65162267 | 0.25 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr6_+_67873135 | 0.25 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr9_+_62249730 | 0.24 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr15_-_99149810 | 0.24 |
ENSMUST00000163506.3
ENSMUST00000229671.2 ENSMUST00000229359.2 ENSMUST00000041190.17 |
Mcrs1
|
microspherule protein 1 |
| chr5_-_92231517 | 0.24 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_-_107925159 | 0.23 |
ENSMUST00000004140.11
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr10_+_43097460 | 0.23 |
ENSMUST00000095725.11
|
Pdss2
|
prenyl (solanesyl) diphosphate synthase, subunit 2 |
| chr5_-_24782465 | 0.20 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr9_-_59393893 | 0.20 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr2_+_5956251 | 0.20 |
ENSMUST00000060092.13
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr9_+_108782646 | 0.20 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr9_+_37466989 | 0.19 |
ENSMUST00000213126.2
|
Siae
|
sialic acid acetylesterase |
| chr12_-_15866763 | 0.19 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr16_+_91203123 | 0.17 |
ENSMUST00000023691.12
|
Il10rb
|
interleukin 10 receptor, beta |
| chr1_+_74627445 | 0.17 |
ENSMUST00000113733.10
ENSMUST00000027358.11 |
Bcs1l
|
BCS1-like (yeast) |
| chr2_-_152673032 | 0.16 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr18_-_10610048 | 0.16 |
ENSMUST00000115864.8
ENSMUST00000145320.2 ENSMUST00000097670.10 |
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr4_+_152423075 | 0.15 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr13_-_35211060 | 0.15 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chrX_-_93166964 | 0.15 |
ENSMUST00000137853.8
|
Zfx
|
zinc finger protein X-linked |
| chr2_+_32496990 | 0.14 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr9_+_108782664 | 0.14 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr1_-_128030148 | 0.14 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr11_-_102298748 | 0.14 |
ENSMUST00000124755.2
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr12_+_99850756 | 0.13 |
ENSMUST00000153627.8
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr9_+_75532992 | 0.12 |
ENSMUST00000034702.6
|
Lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr9_+_62249341 | 0.12 |
ENSMUST00000135395.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr16_+_31918599 | 0.12 |
ENSMUST00000115168.9
|
Cep19
|
centrosomal protein 19 |
| chr11_-_119910986 | 0.11 |
ENSMUST00000134319.2
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr5_-_34345014 | 0.11 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr17_+_84990541 | 0.11 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr4_+_129030710 | 0.10 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr19_-_5538370 | 0.09 |
ENSMUST00000124334.8
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr7_+_45434755 | 0.08 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr17_+_34812361 | 0.08 |
ENSMUST00000174532.2
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr1_-_120001752 | 0.07 |
ENSMUST00000056089.8
|
Tmem37
|
transmembrane protein 37 |
| chr5_-_120605361 | 0.07 |
ENSMUST00000132916.2
|
Sdsl
|
serine dehydratase-like |
| chr9_+_72714156 | 0.06 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr6_+_42263609 | 0.06 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr16_-_31918485 | 0.06 |
ENSMUST00000189013.8
ENSMUST00000155966.8 ENSMUST00000096109.11 ENSMUST00000232321.2 |
Pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr17_+_79934096 | 0.06 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr12_-_114419703 | 0.06 |
ENSMUST00000193397.2
|
Ighv6-7
|
immunoglobulin heavy variable V6-7 |
| chr6_+_67243967 | 0.04 |
ENSMUST00000203436.3
ENSMUST00000205106.3 ENSMUST00000204293.3 ENSMUST00000203077.3 ENSMUST00000204294.3 |
Serbp1
|
serpine1 mRNA binding protein 1 |
| chr6_+_119456629 | 0.04 |
ENSMUST00000032094.7
|
Fbxl14
|
F-box and leucine-rich repeat protein 14 |
| chr15_-_99149794 | 0.04 |
ENSMUST00000229926.2
|
Mcrs1
|
microspherule protein 1 |
| chr10_-_41592630 | 0.04 |
ENSMUST00000189488.3
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr8_-_88686188 | 0.04 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr6_+_87755046 | 0.04 |
ENSMUST00000032133.5
|
Gp9
|
glycoprotein 9 (platelet) |
| chrX_+_162694397 | 0.04 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_-_60667161 | 0.04 |
ENSMUST00000218637.2
|
Unc5b
|
unc-5 netrin receptor B |
| chr4_-_3185358 | 0.03 |
ENSMUST00000105159.5
|
Vmn1r3
|
vomeronasal 1 receptor 3 |
| chr13_+_74498430 | 0.03 |
ENSMUST00000160021.8
ENSMUST00000162672.8 ENSMUST00000162376.2 |
Ccdc127
|
coiled-coil domain containing 127 |
| chr11_+_87590720 | 0.03 |
ENSMUST00000040089.5
|
Rnf43
|
ring finger protein 43 |
| chr18_+_35987733 | 0.03 |
ENSMUST00000235337.2
|
Cxxc5
|
CXXC finger 5 |
| chr2_-_88885768 | 0.03 |
ENSMUST00000099798.2
|
Olfr1218
|
olfactory receptor 1218 |
| chr6_-_92458324 | 0.03 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr5_-_143255713 | 0.03 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr2_-_65397809 | 0.03 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr17_+_46608333 | 0.03 |
ENSMUST00000188223.7
ENSMUST00000061722.13 ENSMUST00000166280.8 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr9_-_52591030 | 0.03 |
ENSMUST00000213937.2
|
AI593442
|
expressed sequence AI593442 |
| chr4_+_155334417 | 0.02 |
ENSMUST00000178473.8
ENSMUST00000105627.8 ENSMUST00000097747.9 |
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr2_-_162929732 | 0.02 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr10_+_85707687 | 0.02 |
ENSMUST00000001836.11
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr15_-_37792635 | 0.02 |
ENSMUST00000090150.11
ENSMUST00000150453.2 |
Ncald
|
neurocalcin delta |
| chr9_+_113886208 | 0.02 |
ENSMUST00000135338.3
|
Susd5
|
sushi domain containing 5 |
| chr15_-_102274781 | 0.02 |
ENSMUST00000078508.7
|
Sp7
|
Sp7 transcription factor 7 |
| chr11_+_4207557 | 0.02 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr2_+_85865109 | 0.02 |
ENSMUST00000213865.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr11_+_49355873 | 0.02 |
ENSMUST00000213674.2
ENSMUST00000204518.3 |
Olfr1386
|
olfactory receptor 1386 |
| chr9_-_117080869 | 0.02 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr3_-_96634752 | 0.02 |
ENSMUST00000154679.8
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr6_+_42263644 | 0.01 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr19_-_13796811 | 0.01 |
ENSMUST00000216659.2
|
Olfr1499
|
olfactory receptor 1499 |
| chr4_+_33132502 | 0.01 |
ENSMUST00000029947.6
|
Gabrr1
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 1 |
| chr13_-_99121070 | 0.01 |
ENSMUST00000054425.7
|
H2bl1
|
H2B.L histone variant 1 |
| chr2_-_164285097 | 0.01 |
ENSMUST00000017153.4
|
Sdc4
|
syndecan 4 |
| chr4_+_118522716 | 0.01 |
ENSMUST00000102666.5
|
Olfr62
|
olfactory receptor 62 |
| chrX_+_139243012 | 0.01 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr9_+_65536892 | 0.01 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr2_+_85551751 | 0.01 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr13_-_21823691 | 0.01 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr5_-_136911969 | 0.01 |
ENSMUST00000057497.13
ENSMUST00000111103.2 |
Col26a1
|
collagen, type XXVI, alpha 1 |
| chr10_+_38841511 | 0.01 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr17_-_29768531 | 0.01 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chr5_+_35198853 | 0.01 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr18_+_35987791 | 0.01 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr18_-_38734389 | 0.01 |
ENSMUST00000025295.8
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr4_-_133930315 | 0.01 |
ENSMUST00000097849.3
|
Zpld2
|
zona pellucida like domain containing 2 |
| chr9_-_58444754 | 0.01 |
ENSMUST00000213722.2
|
Cd276
|
CD276 antigen |
| chr14_+_32321824 | 0.01 |
ENSMUST00000068938.7
ENSMUST00000228878.2 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr1_+_135980639 | 0.01 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr7_-_30062197 | 0.01 |
ENSMUST00000046351.7
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr16_-_34083200 | 0.00 |
ENSMUST00000114947.2
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr6_+_124689241 | 0.00 |
ENSMUST00000004375.16
|
Phb2
|
prohibitin 2 |
| chr12_-_103641723 | 0.00 |
ENSMUST00000095451.2
|
Serpina16
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
| chr2_-_9888804 | 0.00 |
ENSMUST00000114915.3
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr10_+_19588318 | 0.00 |
ENSMUST00000020185.5
|
Il20ra
|
interleukin 20 receptor, alpha |
| chr14_-_20319242 | 0.00 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr13_-_95359543 | 0.00 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chrX_+_119199956 | 0.00 |
ENSMUST00000113364.10
ENSMUST00000050239.16 ENSMUST00000113358.10 |
Pcdh11x
|
protocadherin 11 X-linked |
| chr15_-_102275403 | 0.00 |
ENSMUST00000229464.2
|
Sp7
|
Sp7 transcription factor 7 |
| chr10_+_128212953 | 0.00 |
ENSMUST00000014642.10
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr7_+_12246415 | 0.00 |
ENSMUST00000032541.5
|
2900092C05Rik
|
RIKEN cDNA 2900092C05 gene |
| chr7_-_118594365 | 0.00 |
ENSMUST00000008878.10
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr15_-_78090591 | 0.00 |
ENSMUST00000120592.2
|
Pvalb
|
parvalbumin |
| chr11_-_33793461 | 0.00 |
ENSMUST00000101368.9
ENSMUST00000065970.6 ENSMUST00000109340.9 |
Kcnip1
|
Kv channel-interacting protein 1 |
| chr3_+_82915031 | 0.00 |
ENSMUST00000048486.13
ENSMUST00000194175.2 |
Fgg
|
fibrinogen gamma chain |
| chr5_-_87686048 | 0.00 |
ENSMUST00000031199.11
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr4_+_47288287 | 0.00 |
ENSMUST00000146967.2
|
Col15a1
|
collagen, type XV, alpha 1 |
| chr6_+_4504814 | 0.00 |
ENSMUST00000141483.8
|
Col1a2
|
collagen, type I, alpha 2 |
| chr13_+_3887757 | 0.00 |
ENSMUST00000042219.6
|
Calm4
|
calmodulin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Plagl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 0.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 0.8 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.2 | 0.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 1.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.4 | GO:0044206 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.1 | 0.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.5 | GO:0071931 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.3 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.7 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 1.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.8 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 1.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.6 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 1.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 1.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 0.9 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 1.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 0.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 0.9 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.3 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.8 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0030546 | receptor activator activity(GO:0030546) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |