Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
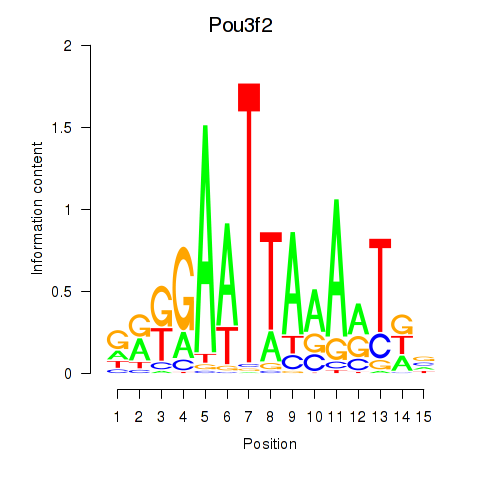
Results for Pou3f2
Z-value: 1.54
Transcription factors associated with Pou3f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f2
|
ENSMUSG00000095139.3 | POU domain, class 3, transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f2 | mm39_v1_chr4_-_22488296_22488366 | 0.96 | 8.5e-03 | Click! |
Activity profile of Pou3f2 motif
Sorted Z-values of Pou3f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_88947627 | 2.57 |
ENSMUST00000217635.2
ENSMUST00000143255.3 ENSMUST00000213404.2 |
Olfr1221
|
olfactory receptor 1221 |
| chr1_+_173983199 | 2.03 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr5_+_107655487 | 1.94 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr13_-_110416637 | 1.19 |
ENSMUST00000167824.3
ENSMUST00000224180.2 |
Rab3c
|
RAB3C, member RAS oncogene family |
| chr2_+_88470886 | 1.04 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr7_-_85525474 | 1.03 |
ENSMUST00000077478.8
ENSMUST00000233729.2 ENSMUST00000233079.2 ENSMUST00000233315.2 ENSMUST00000233115.2 ENSMUST00000233748.2 ENSMUST00000232965.2 ENSMUST00000232897.2 |
Vmn2r73
|
vomeronasal 2, receptor 73 |
| chr10_+_17598961 | 0.95 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr9_+_44893077 | 0.89 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr10_+_129539079 | 0.86 |
ENSMUST00000213331.2
|
Olfr804
|
olfactory receptor 804 |
| chr9_+_48073296 | 0.86 |
ENSMUST00000216998.2
ENSMUST00000215780.2 |
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr1_-_150341911 | 0.83 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr14_-_104081119 | 0.79 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr2_+_134627987 | 0.77 |
ENSMUST00000131552.5
ENSMUST00000110116.8 |
Plcb1
|
phospholipase C, beta 1 |
| chrX_-_125723491 | 0.75 |
ENSMUST00000081074.5
|
4932411N23Rik
|
RIKEN cDNA 4932411N23 gene |
| chr2_-_144369261 | 0.75 |
ENSMUST00000163701.2
ENSMUST00000081982.12 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr10_-_129524028 | 0.70 |
ENSMUST00000203785.3
ENSMUST00000217576.2 |
Olfr802
|
olfactory receptor 802 |
| chr17_-_37523969 | 0.70 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr1_+_9978863 | 0.64 |
ENSMUST00000052843.12
ENSMUST00000171802.8 ENSMUST00000125294.9 ENSMUST00000140948.9 |
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr4_-_108637700 | 0.63 |
ENSMUST00000106658.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr2_-_88810595 | 0.62 |
ENSMUST00000168169.10
ENSMUST00000144908.3 |
Olfr1213
|
olfactory receptor 1213 |
| chr7_+_75259778 | 0.61 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr10_-_130002635 | 0.60 |
ENSMUST00000216530.2
|
Olfr825
|
olfactory receptor 825 |
| chr5_-_66308421 | 0.60 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr11_+_60244132 | 0.60 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr3_+_137573839 | 0.57 |
ENSMUST00000197711.2
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr7_-_80051455 | 0.54 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr4_-_108637979 | 0.53 |
ENSMUST00000106657.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr7_-_12829100 | 0.52 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr2_-_89030312 | 0.51 |
ENSMUST00000214709.2
ENSMUST00000215987.2 ENSMUST00000215562.2 ENSMUST00000220416.2 ENSMUST00000216445.2 ENSMUST00000217601.2 |
Olfr1226
|
olfactory receptor 1226 |
| chr16_+_44687460 | 0.50 |
ENSMUST00000102805.4
|
Cd200r2
|
Cd200 receptor 2 |
| chr10_+_57670431 | 0.50 |
ENSMUST00000151623.8
ENSMUST00000020022.8 |
Smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr1_+_150268544 | 0.47 |
ENSMUST00000124973.9
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr6_+_90078412 | 0.45 |
ENSMUST00000089417.8
ENSMUST00000226577.2 |
Vmn1r50
|
vomeronasal 1 receptor 50 |
| chr16_-_36695166 | 0.45 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chr17_-_37627945 | 0.44 |
ENSMUST00000217590.2
|
Olfr102
|
olfactory receptor 102 |
| chr3_-_27765381 | 0.42 |
ENSMUST00000193779.2
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr8_+_85897951 | 0.42 |
ENSMUST00000076896.6
|
Cks1brt
|
CDC28 protein kinase 1b, retrogene |
| chr2_+_88479038 | 0.42 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1 |
| chr9_-_88974735 | 0.41 |
ENSMUST00000189557.2
ENSMUST00000167113.8 |
Trim43b
|
tripartite motif-containing 43B |
| chr2_-_87838612 | 0.41 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr6_-_119940694 | 0.40 |
ENSMUST00000161512.3
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr1_-_155912159 | 0.40 |
ENSMUST00000097527.10
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr17_-_45047521 | 0.40 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr7_-_140402037 | 0.39 |
ENSMUST00000106052.2
ENSMUST00000080651.13 |
Zfp941
|
zinc finger protein 941 |
| chr11_-_60243695 | 0.37 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr15_-_83033471 | 0.37 |
ENSMUST00000129372.2
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr2_+_111120973 | 0.37 |
ENSMUST00000213727.2
|
Olfr1278
|
olfactory receptor 1278 |
| chr14_-_104081827 | 0.34 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr13_+_25240138 | 0.34 |
ENSMUST00000069614.7
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr2_+_61423421 | 0.34 |
ENSMUST00000112495.8
ENSMUST00000112501.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr8_-_84963653 | 0.33 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr10_+_117465397 | 0.32 |
ENSMUST00000020399.6
|
Cpm
|
carboxypeptidase M |
| chr3_+_84573499 | 0.32 |
ENSMUST00000107682.2
|
Tmem154
|
transmembrane protein 154 |
| chr17_+_43700327 | 0.31 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr14_+_84680993 | 0.30 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr3_+_66892979 | 0.29 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr6_-_66787481 | 0.29 |
ENSMUST00000228862.2
ENSMUST00000227285.2 ENSMUST00000228008.2 ENSMUST00000228651.2 ENSMUST00000227555.2 |
Vmn1r39
|
vomeronasal 1 receptor 39 |
| chr12_-_91351177 | 0.27 |
ENSMUST00000141429.8
|
Cep128
|
centrosomal protein 128 |
| chr9_+_88721217 | 0.26 |
ENSMUST00000163255.9
ENSMUST00000186363.2 |
Trim43c
|
tripartite motif-containing 43C |
| chr18_+_4920513 | 0.26 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr19_-_6910195 | 0.25 |
ENSMUST00000236443.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr2_-_87543523 | 0.25 |
ENSMUST00000214209.2
|
Olfr1137
|
olfactory receptor 1137 |
| chr13_-_110417421 | 0.24 |
ENSMUST00000223922.2
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr14_+_60615128 | 0.24 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr6_+_13069757 | 0.24 |
ENSMUST00000124234.8
ENSMUST00000142211.8 ENSMUST00000031556.14 |
Tmem106b
|
transmembrane protein 106B |
| chr7_-_29643172 | 0.23 |
ENSMUST00000108212.8
|
Zfp74
|
zinc finger protein 74 |
| chr19_-_7183626 | 0.22 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr9_-_69945358 | 0.22 |
ENSMUST00000034751.6
|
Gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr12_+_52746158 | 0.21 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr5_-_66211842 | 0.21 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr9_+_38278192 | 0.21 |
ENSMUST00000216168.2
|
Olfr250
|
olfactory receptor 250 |
| chr10_+_62860291 | 0.21 |
ENSMUST00000020262.5
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr14_+_73376192 | 0.20 |
ENSMUST00000171070.8
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr1_-_155912216 | 0.20 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr5_-_53864595 | 0.20 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr11_-_99134885 | 0.19 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr10_-_129751466 | 0.19 |
ENSMUST00000213438.2
|
Olfr816
|
olfactory receptor 816 |
| chr12_+_69288606 | 0.19 |
ENSMUST00000063445.13
|
Klhdc1
|
kelch domain containing 1 |
| chr7_+_44485704 | 0.18 |
ENSMUST00000033015.8
|
Il4i1
|
interleukin 4 induced 1 |
| chr15_+_36179676 | 0.18 |
ENSMUST00000171205.3
|
Spag1
|
sperm associated antigen 1 |
| chr17_-_38399584 | 0.17 |
ENSMUST00000216523.2
ENSMUST00000215940.2 ENSMUST00000172933.2 |
Olfr131
|
olfactory receptor 131 |
| chr7_+_104226521 | 0.17 |
ENSMUST00000217466.2
ENSMUST00000215585.3 |
Olfr653
|
olfactory receptor 653 |
| chr17_+_34311314 | 0.17 |
ENSMUST00000025192.8
|
H2-Oa
|
histocompatibility 2, O region alpha locus |
| chr7_+_11905206 | 0.15 |
ENSMUST00000226953.2
ENSMUST00000227530.2 |
Vmn1r79
|
vomeronasal 1 receptor 79 |
| chr4_-_149211145 | 0.15 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chrX_-_58211440 | 0.14 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr2_-_87985537 | 0.13 |
ENSMUST00000216951.2
|
Olfr1167
|
olfactory receptor 1167 |
| chr16_+_36695479 | 0.12 |
ENSMUST00000023534.7
ENSMUST00000114812.9 ENSMUST00000134616.8 |
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr10_-_129630735 | 0.12 |
ENSMUST00000217283.2
ENSMUST00000214206.2 ENSMUST00000214878.2 |
Olfr810
|
olfactory receptor 810 |
| chr19_-_6593049 | 0.12 |
ENSMUST00000113451.9
|
Slc22a12
|
solute carrier family 22 (organic anion/cation transporter), member 12 |
| chr2_-_88837317 | 0.12 |
ENSMUST00000216271.3
ENSMUST00000214809.3 |
Olfr1215
|
olfactory receptor 1215 |
| chr5_+_107645626 | 0.10 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr1_-_135513083 | 0.10 |
ENSMUST00000040599.15
|
Nav1
|
neuron navigator 1 |
| chr2_-_59955995 | 0.10 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_-_134731516 | 0.09 |
ENSMUST00000238280.2
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_-_54655079 | 0.09 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr1_-_135513443 | 0.09 |
ENSMUST00000067414.13
|
Nav1
|
neuron navigator 1 |
| chr8_-_25066313 | 0.09 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr17_+_8164416 | 0.08 |
ENSMUST00000232223.2
ENSMUST00000097423.3 |
Rsph3a
|
radial spoke 3A homolog (Chlamydomonas) |
| chr6_-_12749192 | 0.08 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr2_+_154855451 | 0.08 |
ENSMUST00000029123.3
|
a
|
nonagouti |
| chrX_+_82898974 | 0.08 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chr13_+_83723255 | 0.07 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_72580823 | 0.06 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr11_+_49169131 | 0.05 |
ENSMUST00000213323.2
|
Olfr1393
|
olfactory receptor 1393 |
| chr11_+_73673790 | 0.05 |
ENSMUST00000120081.3
ENSMUST00000215161.2 ENSMUST00000206815.2 |
Olfr390
|
olfactory receptor 390 |
| chr18_-_36828978 | 0.04 |
ENSMUST00000115682.2
|
E230025N22Rik
|
Riken cDNA E230025N22 gene |
| chr6_+_83985684 | 0.04 |
ENSMUST00000203803.3
ENSMUST00000204591.3 ENSMUST00000113823.8 ENSMUST00000153860.4 |
Dysf
|
dysferlin |
| chr11_-_78313043 | 0.04 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr13_+_49736219 | 0.04 |
ENSMUST00000065494.8
|
Omd
|
osteomodulin |
| chr5_+_107579407 | 0.04 |
ENSMUST00000166599.2
|
Lpcat2b
|
lysophosphatidylcholine acyltransferase 2B |
| chr2_-_89084074 | 0.03 |
ENSMUST00000216392.2
|
Olfr1228
|
olfactory receptor 1228 |
| chr15_-_65848649 | 0.03 |
ENSMUST00000239034.2
ENSMUST00000100584.3 |
Hhla1
|
HERV-H LTR-associating 1 |
| chr7_+_30399208 | 0.03 |
ENSMUST00000013227.8
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr9_+_88462944 | 0.03 |
ENSMUST00000164661.4
ENSMUST00000215498.2 ENSMUST00000216686.2 |
Trim43a
|
tripartite motif-containing 43A |
| chr3_+_108646974 | 0.02 |
ENSMUST00000133931.9
|
Aknad1
|
AKNA domain containing 1 |
| chr6_-_129717431 | 0.02 |
ENSMUST00000050385.6
|
Klri2
|
killer cell lectin-like receptor family I member 2 |
| chr6_-_69245427 | 0.02 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr3_+_28038283 | 0.02 |
ENSMUST00000067757.11
ENSMUST00000123539.8 |
Pld1
|
phospholipase D1 |
| chr19_+_31060237 | 0.02 |
ENSMUST00000066039.8
|
Cstf2t
|
cleavage stimulation factor, 3' pre-RNA subunit 2, tau |
| chr9_+_37656402 | 0.02 |
ENSMUST00000216982.2
|
Olfr874
|
olfactory receptor 874 |
| chr13_-_23041731 | 0.02 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr13_-_51723473 | 0.01 |
ENSMUST00000239056.2
ENSMUST00000223543.3 |
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr13_+_55932387 | 0.01 |
ENSMUST00000109898.3
|
Catsper3
|
cation channel, sperm associated 3 |
| chr7_-_132616977 | 0.01 |
ENSMUST00000169570.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr3_+_55369149 | 0.01 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr16_-_92622659 | 0.01 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr8_+_21805562 | 0.01 |
ENSMUST00000167683.3
ENSMUST00000168340.2 |
Defa27
|
defensin, alpha, 27 |
| chr6_+_34006356 | 0.01 |
ENSMUST00000228187.2
ENSMUST00000070189.10 ENSMUST00000101564.9 |
Lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr6_-_69394425 | 0.01 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr13_+_55932369 | 0.01 |
ENSMUST00000021961.12
|
Catsper3
|
cation channel, sperm associated 3 |
| chr9_+_54441404 | 0.01 |
ENSMUST00000118413.3
|
Sh2d7
|
SH2 domain containing 7 |
| chr17_+_24026892 | 0.01 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr1_-_162641495 | 0.01 |
ENSMUST00000144916.8
ENSMUST00000140274.2 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr13_-_64645606 | 0.00 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr19_-_20368029 | 0.00 |
ENSMUST00000235280.2
|
Anxa1
|
annexin A1 |
| chr6_-_40917431 | 0.00 |
ENSMUST00000122181.8
ENSMUST00000031935.10 |
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr11_-_23469181 | 0.00 |
ENSMUST00000239488.2
ENSMUST00000020527.13 |
1700093K21Rik
|
RIKEN cDNA 1700093K21 gene |
| chr5_-_24732200 | 0.00 |
ENSMUST00000088311.6
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr2_-_136229849 | 0.00 |
ENSMUST00000035264.9
ENSMUST00000077200.4 |
Pak5
|
p21 (RAC1) activated kinase 5 |
| chr7_-_20832438 | 0.00 |
ENSMUST00000105197.3
|
Vmn1r121
|
vomeronasal 1 receptor 121 |
| chrX_-_52778353 | 0.00 |
ENSMUST00000071711.4
|
Zfp36l3
|
zinc finger protein 36, C3H type-like 3 |
| chr8_+_110717062 | 0.00 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chrX_-_93408176 | 0.00 |
ENSMUST00000239046.2
|
CXorf58
|
family with sequence similarity 90, member A1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1904117 | interleukin-12-mediated signaling pathway(GO:0035722) activation of meiosis involved in egg activation(GO:0060466) cellular response to interleukin-12(GO:0071349) response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.2 | 1.0 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.2 | 0.6 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 1.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.2 | 0.5 | GO:0006404 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.6 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.3 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.9 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.2 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.4 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 6.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 1.4 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |