Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Pou3f3
Z-value: 0.46
Transcription factors associated with Pou3f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f3
|
ENSMUSG00000045515.5 | POU domain, class 3, transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f3 | mm39_v1_chr1_+_42734051_42734105 | -1.00 | 3.6e-04 | Click! |
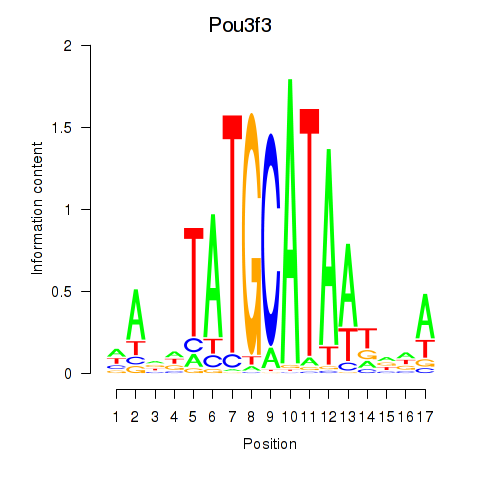
Activity profile of Pou3f3 motif
Sorted Z-values of Pou3f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_129326927 | 0.27 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr4_+_134238310 | 0.27 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr7_-_126391388 | 0.23 |
ENSMUST00000206570.2
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr15_-_42540363 | 0.20 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr7_-_126391657 | 0.19 |
ENSMUST00000032936.8
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr14_+_26722319 | 0.19 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr4_-_149222057 | 0.17 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr10_+_26105605 | 0.16 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr4_-_149221998 | 0.13 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr6_+_54249817 | 0.11 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr2_-_30249202 | 0.11 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr1_+_43571536 | 0.10 |
ENSMUST00000202540.2
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chrX_+_149377416 | 0.07 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr8_-_106022676 | 0.06 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr7_+_19741948 | 0.03 |
ENSMUST00000073151.13
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr3_-_144555062 | 0.02 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr2_+_110551927 | 0.02 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr3_-_122828592 | 0.02 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr2_+_67004178 | 0.02 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr2_-_88981904 | 0.02 |
ENSMUST00000217342.2
|
Olfr1223
|
olfactory receptor 1223 |
| chr2_+_110551976 | 0.02 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr5_+_66833434 | 0.01 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr9_+_50466127 | 0.01 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr11_+_9141934 | 0.01 |
ENSMUST00000042740.13
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr7_+_20521210 | 0.01 |
ENSMUST00000173723.2
ENSMUST00000168794.2 |
Vmn1r113
|
vomeronasal 1 receptor 113 |
| chr6_+_58242159 | 0.01 |
ENSMUST00000228678.2
|
Vmn1r28
|
vomeronasal 1 receptor 28 |
| chr6_+_42932393 | 0.01 |
ENSMUST00000095955.3
|
Olfr444
|
olfactory receptor 444 |
| chr3_+_92223927 | 0.01 |
ENSMUST00000061038.4
|
Sprr2b
|
small proline-rich protein 2B |
| chr2_-_36818909 | 0.01 |
ENSMUST00000213574.2
|
Olfr355
|
olfactory receptor 355 |
| chr6_-_101176147 | 0.01 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr2_-_88818544 | 0.01 |
ENSMUST00000099804.2
|
Olfr1214
|
olfactory receptor 1214 |
| chr3_+_83673606 | 0.01 |
ENSMUST00000029625.8
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr7_-_106408041 | 0.01 |
ENSMUST00000214840.2
|
Olfr700
|
olfactory receptor 700 |
| chr15_-_83949528 | 0.01 |
ENSMUST00000156187.8
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr2_+_86828742 | 0.01 |
ENSMUST00000214002.2
|
Olfr1102
|
olfactory receptor 1102 |
| chr3_-_145813802 | 0.01 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3 |
| chr5_-_66672158 | 0.00 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr7_-_6489812 | 0.00 |
ENSMUST00000086318.5
ENSMUST00000209866.4 |
Olfr5
|
olfactory receptor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |