Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
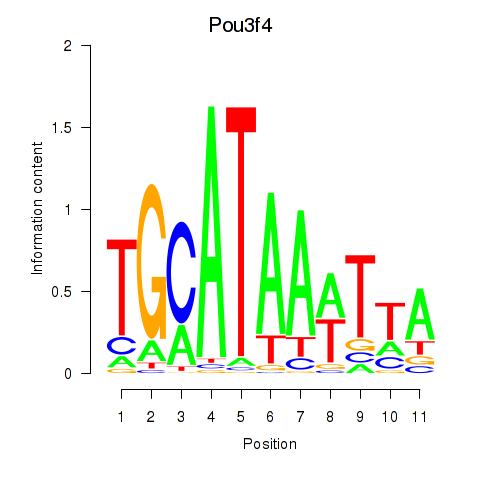
Results for Pou3f4
Z-value: 0.62
Transcription factors associated with Pou3f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f4
|
ENSMUSG00000056854.5 | POU domain, class 3, transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f4 | mm39_v1_chrX_+_109857866_109857886 | -0.88 | 5.2e-02 | Click! |
Activity profile of Pou3f4 motif
Sorted Z-values of Pou3f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_60907698 | 0.90 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr18_+_60907668 | 0.72 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr3_+_102641822 | 0.58 |
ENSMUST00000029451.12
|
Tspan2
|
tetraspanin 2 |
| chr9_+_113641615 | 0.37 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr6_-_56681657 | 0.35 |
ENSMUST00000176595.3
ENSMUST00000170382.5 ENSMUST00000203958.2 |
Lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_-_105398346 | 0.33 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr10_+_7543260 | 0.33 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr11_+_29668563 | 0.32 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr11_-_101357046 | 0.31 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chr9_+_48966868 | 0.31 |
ENSMUST00000034803.10
|
Zw10
|
zw10 kinetochore protein |
| chr1_+_179788037 | 0.30 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_-_126391388 | 0.27 |
ENSMUST00000206570.2
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr15_+_84076423 | 0.25 |
ENSMUST00000023071.8
|
Samm50
|
SAMM50 sorting and assembly machinery component |
| chr10_+_79650496 | 0.24 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr4_+_152123772 | 0.24 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr9_-_55419442 | 0.24 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr4_+_6365650 | 0.23 |
ENSMUST00000029912.11
ENSMUST00000103008.12 |
Sdcbp
|
syndecan binding protein |
| chr17_+_87270504 | 0.23 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr3_+_94600863 | 0.23 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr2_-_125701059 | 0.23 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr2_+_173579285 | 0.22 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr7_+_101872227 | 0.22 |
ENSMUST00000156529.8
ENSMUST00000138479.8 ENSMUST00000126914.8 ENSMUST00000033292.14 ENSMUST00000146559.2 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr1_-_165762469 | 0.21 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr18_+_56565188 | 0.20 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr16_-_22258469 | 0.20 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr2_+_125701054 | 0.20 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr7_-_126391657 | 0.20 |
ENSMUST00000032936.8
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr11_-_79853200 | 0.20 |
ENSMUST00000108241.8
ENSMUST00000043152.6 |
Utp6
|
UTP6 small subunit processome component |
| chr1_+_179788675 | 0.19 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr11_-_74615496 | 0.19 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr4_+_6365694 | 0.19 |
ENSMUST00000175769.8
ENSMUST00000140830.8 ENSMUST00000108374.8 |
Sdcbp
|
syndecan binding protein |
| chr8_-_61407760 | 0.19 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr17_-_22127073 | 0.18 |
ENSMUST00000106026.3
|
Zfp995
|
zinc finger protein 995 |
| chr13_+_93441307 | 0.17 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chrX_+_149377416 | 0.17 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_-_52827015 | 0.17 |
ENSMUST00000031069.13
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr2_-_30857858 | 0.17 |
ENSMUST00000028200.9
|
Tor1a
|
torsin family 1, member A (torsin A) |
| chr11_+_93935021 | 0.16 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr3_+_85946145 | 0.16 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr4_-_138053545 | 0.16 |
ENSMUST00000105817.4
|
Pink1
|
PTEN induced putative kinase 1 |
| chr2_+_30254239 | 0.16 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chrX_+_20529137 | 0.16 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr10_-_57408512 | 0.16 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr11_+_93935066 | 0.15 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr15_-_98729333 | 0.14 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr6_+_145067457 | 0.14 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr13_-_62519750 | 0.14 |
ENSMUST00000107989.7
|
Gm3604
|
predicted gene 3604 |
| chr6_-_134864731 | 0.14 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr9_+_96141299 | 0.13 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr1_+_43571536 | 0.13 |
ENSMUST00000202540.2
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr9_+_96141317 | 0.13 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr5_+_38233901 | 0.12 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr7_+_101871623 | 0.12 |
ENSMUST00000143541.8
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr11_+_93935156 | 0.12 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr18_-_75094323 | 0.12 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr17_+_87270707 | 0.11 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr5_-_146157711 | 0.11 |
ENSMUST00000169407.9
ENSMUST00000161331.8 ENSMUST00000159074.3 ENSMUST00000067837.10 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr9_+_14695933 | 0.11 |
ENSMUST00000034405.11
ENSMUST00000115632.10 ENSMUST00000147305.2 |
Mre11a
|
MRE11A homolog A, double strand break repair nuclease |
| chr3_+_67799510 | 0.11 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr5_-_92583078 | 0.11 |
ENSMUST00000038514.15
|
Nup54
|
nucleoporin 54 |
| chr7_-_90125178 | 0.11 |
ENSMUST00000032843.9
|
Tmem126b
|
transmembrane protein 126B |
| chr9_+_96140781 | 0.10 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chrX_+_152506577 | 0.10 |
ENSMUST00000140575.8
ENSMUST00000208373.2 ENSMUST00000185492.7 ENSMUST00000149514.8 |
Nbdy
|
negative regulator of P-body association |
| chr19_-_32085532 | 0.10 |
ENSMUST00000236030.2
ENSMUST00000237104.2 ENSMUST00000236208.2 ENSMUST00000235273.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr3_+_105881027 | 0.10 |
ENSMUST00000000573.9
|
Ovgp1
|
oviductal glycoprotein 1 |
| chr11_+_93934940 | 0.10 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr3_+_68479578 | 0.10 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr13_+_93440572 | 0.09 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr16_+_16688692 | 0.09 |
ENSMUST00000232547.2
|
Top3b
|
topoisomerase (DNA) III beta |
| chr8_-_72178340 | 0.09 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr13_+_104424359 | 0.09 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr7_+_101530524 | 0.09 |
ENSMUST00000106978.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr15_-_100497863 | 0.08 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr9_+_19533374 | 0.08 |
ENSMUST00000213725.2
ENSMUST00000208694.2 |
Zfp317
|
zinc finger protein 317 |
| chr4_+_155048571 | 0.07 |
ENSMUST00000030931.11
ENSMUST00000070953.11 |
Pank4
|
pantothenate kinase 4 |
| chr3_+_94840352 | 0.07 |
ENSMUST00000090839.12
|
Selenbp1
|
selenium binding protein 1 |
| chr3_-_51184730 | 0.06 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr15_-_9140403 | 0.06 |
ENSMUST00000096482.10
|
Skp2
|
S-phase kinase-associated protein 2 |
| chr8_+_96551974 | 0.06 |
ENSMUST00000074053.6
|
Sap18b
|
Sin3-associated polypeptide 18B |
| chr9_-_40257586 | 0.06 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr7_-_66915756 | 0.06 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr9_+_19533591 | 0.06 |
ENSMUST00000215372.2
|
Zfp317
|
zinc finger protein 317 |
| chr11_-_100830183 | 0.05 |
ENSMUST00000092671.12
ENSMUST00000103114.8 |
Stat3
|
signal transducer and activator of transcription 3 |
| chr3_-_51184895 | 0.05 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr12_-_25147139 | 0.05 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr10_-_128759817 | 0.05 |
ENSMUST00000131271.2
|
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr12_-_59108211 | 0.05 |
ENSMUST00000217863.2
ENSMUST00000021380.10 |
Trappc6b
|
trafficking protein particle complex 6B |
| chr19_+_37425180 | 0.04 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr9_+_96140750 | 0.04 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr18_+_44237474 | 0.04 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr15_-_9140460 | 0.03 |
ENSMUST00000110585.10
|
Skp2
|
S-phase kinase-associated protein 2 |
| chr7_-_100164007 | 0.03 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr2_-_109111064 | 0.03 |
ENSMUST00000147770.2
|
Mettl15
|
methyltransferase like 15 |
| chrX_-_42256694 | 0.03 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr2_+_152873772 | 0.03 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr17_-_59298338 | 0.03 |
ENSMUST00000025064.14
|
Pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr2_+_110551927 | 0.03 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr16_+_94225942 | 0.03 |
ENSMUST00000141176.2
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr1_-_180823709 | 0.02 |
ENSMUST00000154133.8
|
Ephx1
|
epoxide hydrolase 1, microsomal |
| chr2_-_164231015 | 0.02 |
ENSMUST00000167427.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr3_-_116047148 | 0.02 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr5_+_28370687 | 0.02 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chrX_-_87159237 | 0.02 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr9_-_26713874 | 0.02 |
ENSMUST00000162252.8
|
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr2_+_110551976 | 0.02 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr4_-_45532470 | 0.02 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr14_+_53310220 | 0.02 |
ENSMUST00000196079.2
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chr16_-_22258320 | 0.02 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chrX_+_95498965 | 0.02 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr6_+_65502344 | 0.02 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr7_-_106574660 | 0.02 |
ENSMUST00000051715.8
ENSMUST00000207200.3 ENSMUST00000216335.2 |
Olfr711
|
olfactory receptor 711 |
| chr18_+_22478146 | 0.02 |
ENSMUST00000120223.8
ENSMUST00000097655.4 |
Asxl3
|
ASXL transcriptional regulator 3 |
| chr2_+_109111083 | 0.02 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr6_-_113696809 | 0.02 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr2_-_86429310 | 0.02 |
ENSMUST00000215600.2
|
Olfr1082
|
olfactory receptor 1082 |
| chrX_+_162945162 | 0.01 |
ENSMUST00000131543.2
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr2_-_88990751 | 0.01 |
ENSMUST00000216833.2
ENSMUST00000216976.2 |
Olfr1224-ps1
|
olfactory receptor 1224, pseudogene 1 |
| chr12_+_103564479 | 0.01 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr12_-_114321838 | 0.01 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr14_+_27598021 | 0.01 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr3_+_68776884 | 0.01 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr2_+_84966569 | 0.01 |
ENSMUST00000057019.9
|
Aplnr
|
apelin receptor |
| chr3_-_33136153 | 0.01 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr10_+_129336682 | 0.01 |
ENSMUST00000056002.5
ENSMUST00000218901.2 |
Olfr790
|
olfactory receptor 790 |
| chr4_-_110144676 | 0.01 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr8_+_11808349 | 0.01 |
ENSMUST00000074856.13
ENSMUST00000098938.9 |
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr2_+_67004178 | 0.01 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr11_-_100830288 | 0.01 |
ENSMUST00000138438.2
|
Stat3
|
signal transducer and activator of transcription 3 |
| chr14_+_10123804 | 0.01 |
ENSMUST00000022262.6
ENSMUST00000224714.2 |
Fezf2
|
Fez family zinc finger 2 |
| chr16_-_59182579 | 0.01 |
ENSMUST00000055868.4
ENSMUST00000208246.2 |
Olfr209
|
olfactory receptor 209 |
| chr2_+_67276338 | 0.01 |
ENSMUST00000239060.2
ENSMUST00000028410.4 ENSMUST00000112347.8 ENSMUST00000238878.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr7_+_103119084 | 0.01 |
ENSMUST00000213546.3
|
Olfr608
|
olfactory receptor 608 |
| chr10_-_38998272 | 0.01 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr8_+_10299288 | 0.01 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr13_+_21299864 | 0.01 |
ENSMUST00000079050.3
|
Olfr1369-ps1
|
olfactory receptor 1369, pseudogene 1 |
| chr7_+_107413787 | 0.01 |
ENSMUST00000084756.2
|
Olfr467
|
olfactory receptor 467 |
| chr7_+_107994038 | 0.01 |
ENSMUST00000210990.2
|
Olfr495
|
olfactory receptor 495 |
| chr1_+_106866678 | 0.01 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr11_+_71640739 | 0.01 |
ENSMUST00000150531.2
|
Wscd1
|
WSC domain containing 1 |
| chr10_+_87694117 | 0.01 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr17_-_29162794 | 0.01 |
ENSMUST00000232977.2
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr10_+_97482946 | 0.01 |
ENSMUST00000105285.4
|
Epyc
|
epiphycan |
| chr11_-_99313078 | 0.01 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr6_-_101176147 | 0.01 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr16_+_25620652 | 0.01 |
ENSMUST00000115304.8
ENSMUST00000115305.2 ENSMUST00000040231.13 ENSMUST00000115306.8 |
Trp63
|
transformation related protein 63 |
| chr8_-_86091970 | 0.01 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_-_138053603 | 0.01 |
ENSMUST00000030536.13
|
Pink1
|
PTEN induced putative kinase 1 |
| chr13_-_91053440 | 0.01 |
ENSMUST00000109541.4
|
Atp6ap1l
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr1_-_126758369 | 0.01 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr8_-_86091946 | 0.01 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr9_-_19289965 | 0.01 |
ENSMUST00000216839.3
|
Olfr847
|
olfactory receptor 847 |
| chr5_+_57876401 | 0.01 |
ENSMUST00000094783.7
|
Pcdh7
|
protocadherin 7 |
| chr10_-_129784956 | 0.01 |
ENSMUST00000215527.2
ENSMUST00000205227.2 |
Olfr818
|
olfactory receptor 818 |
| chr2_+_86828742 | 0.01 |
ENSMUST00000214002.2
|
Olfr1102
|
olfactory receptor 1102 |
| chr8_+_94537910 | 0.01 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr2_+_111311164 | 0.01 |
ENSMUST00000120021.5
|
Olfr1289
|
olfactory receptor 1289 |
| chr3_-_57483175 | 0.01 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr6_-_130258891 | 0.01 |
ENSMUST00000112020.2
|
Klra10
|
killer cell lectin-like receptor subfamily A, member 10 |
| chr17_-_40553176 | 0.01 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr2_-_88905433 | 0.01 |
ENSMUST00000099797.2
|
Olfr1219
|
olfactory receptor 1219 |
| chr13_-_56399976 | 0.01 |
ENSMUST00000058475.6
|
Neurog1
|
neurogenin 1 |
| chr6_+_66708376 | 0.01 |
ENSMUST00000077482.2
|
Vmn1r37
|
vomeronasal 1 receptor 37 |
| chr11_-_58446443 | 0.01 |
ENSMUST00000216725.2
ENSMUST00000215717.2 ENSMUST00000108824.3 |
Olfr328
|
olfactory receptor 328 |
| chr18_+_37544717 | 0.01 |
ENSMUST00000051126.4
|
Pcdhb10
|
protocadherin beta 10 |
| chr2_+_88012224 | 0.01 |
ENSMUST00000215996.2
|
Olfr1168
|
olfactory receptor 1168 |
| chr10_+_87695117 | 0.01 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr10_-_23869379 | 0.01 |
ENSMUST00000078532.3
|
Taar7a
|
trace amine-associated receptor 7A |
| chr5_-_137784943 | 0.01 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme |
| chr4_+_13784749 | 0.01 |
ENSMUST00000098256.4
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr4_+_52607204 | 0.01 |
ENSMUST00000107671.2
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr2_-_79959802 | 0.01 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr16_-_59104858 | 0.01 |
ENSMUST00000049859.2
|
Olfr202
|
olfactory receptor 202 |
| chr7_+_108064533 | 0.01 |
ENSMUST00000217616.3
|
Olfr498
|
olfactory receptor 498 |
| chr6_+_57090410 | 0.01 |
ENSMUST00000176073.3
|
Vmn1r10
|
vomeronasal 1 receptor 10 |
| chr1_-_25868592 | 0.01 |
ENSMUST00000135518.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr10_-_61814852 | 0.01 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr2_+_88167065 | 0.01 |
ENSMUST00000213778.2
|
Olfr1176
|
olfactory receptor 1176 |
| chr6_+_21215472 | 0.01 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr4_-_36056726 | 0.01 |
ENSMUST00000108124.4
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr7_-_107855343 | 0.01 |
ENSMUST00000072968.2
|
Olfr488
|
olfactory receptor 488 |
| chr1_+_66360865 | 0.00 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr9_+_38358948 | 0.00 |
ENSMUST00000213105.2
|
Olfr902
|
olfactory receptor 902 |
| chr7_+_107964542 | 0.00 |
ENSMUST00000209743.2
|
Olfr494
|
olfactory receptor 494 |
| chr7_-_103289443 | 0.00 |
ENSMUST00000058744.5
|
Olfr622
|
olfactory receptor 622 |
| chr18_+_37637317 | 0.00 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr7_-_101765915 | 0.00 |
ENSMUST00000084830.2
|
Chrna10
|
cholinergic receptor, nicotinic, alpha polypeptide 10 |
| chr10_+_90412114 | 0.00 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_-_126758520 | 0.00 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr3_-_57483330 | 0.00 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr3_-_102074246 | 0.00 |
ENSMUST00000161021.2
|
Vangl1
|
VANGL planar cell polarity 1 |
| chr2_-_66271097 | 0.00 |
ENSMUST00000112371.9
ENSMUST00000138910.3 |
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr19_+_12930229 | 0.00 |
ENSMUST00000213587.2
|
Olfr1450
|
olfactory receptor 1450 |
| chr12_-_113589576 | 0.00 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr7_+_107702486 | 0.00 |
ENSMUST00000104917.4
|
Olfr483
|
olfactory receptor 483 |
| chr11_+_67061837 | 0.00 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr15_+_9140685 | 0.00 |
ENSMUST00000090380.6
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr7_-_103261741 | 0.00 |
ENSMUST00000052152.3
|
Olfr620
|
olfactory receptor 620 |
| chr19_-_12881094 | 0.00 |
ENSMUST00000208343.2
ENSMUST00000216989.2 |
Olfr1447
|
olfactory receptor 1447 |
| chr4_+_15881256 | 0.00 |
ENSMUST00000029876.2
|
Calb1
|
calbindin 1 |
| chrX_+_147454287 | 0.00 |
ENSMUST00000112753.4
|
Gm15127
|
predicted gene 15127 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.2 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.6 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.2 | GO:0046725 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.2 | GO:0051940 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.2 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.3 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.1 | 0.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 1.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0048273 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0070095 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |