Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Pou5f1
Z-value: 1.35
Transcription factors associated with Pou5f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou5f1
|
ENSMUSG00000024406.17 | POU domain, class 5, transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou5f1 | mm39_v1_chr17_+_35816915_35816968 | 0.93 | 2.1e-02 | Click! |
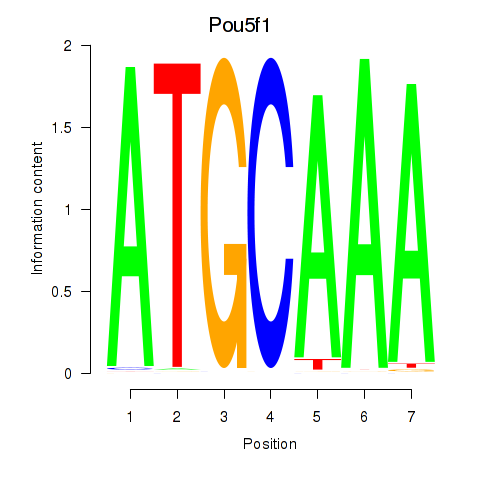
Activity profile of Pou5f1 motif
Sorted Z-values of Pou5f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23930717 | 2.04 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr13_-_23758370 | 1.84 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7 |
| chr3_+_96177010 | 1.69 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr3_-_96128196 | 1.65 |
ENSMUST00000090782.4
|
H2ac20
|
H2A clustered histone 20 |
| chr13_+_22220000 | 1.57 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr13_+_21906214 | 1.49 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr5_+_64969679 | 1.47 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr13_+_22017906 | 1.36 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr11_+_58845502 | 1.31 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr13_+_23868175 | 1.25 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr13_+_22227359 | 1.25 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr13_+_23755551 | 1.11 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr13_-_24118139 | 1.05 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr3_+_96127174 | 0.96 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr13_+_23935088 | 0.95 |
ENSMUST00000078369.3
|
H2ac4
|
H2A clustered histone 4 |
| chr11_-_23720953 | 0.90 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr15_+_6552270 | 0.90 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr13_-_54836059 | 0.89 |
ENSMUST00000122935.2
ENSMUST00000128257.8 |
Rnf44
|
ring finger protein 44 |
| chr8_-_123405392 | 0.83 |
ENSMUST00000134045.2
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr6_-_70237939 | 0.82 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr14_-_67106037 | 0.82 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr16_-_4698148 | 0.78 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr12_-_99378855 | 0.74 |
ENSMUST00000177269.2
|
Foxn3
|
forkhead box N3 |
| chr5_+_129661233 | 0.73 |
ENSMUST00000031390.10
|
Mmp17
|
matrix metallopeptidase 17 |
| chr13_-_23867924 | 0.71 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr6_+_136495818 | 0.70 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr4_+_119671688 | 0.69 |
ENSMUST00000106307.9
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr8_-_116434517 | 0.69 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr9_+_106099797 | 0.69 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr8_+_108669276 | 0.69 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr15_+_6609322 | 0.68 |
ENSMUST00000090461.12
|
Fyb
|
FYN binding protein |
| chr8_+_91635192 | 0.68 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_51076724 | 0.67 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr13_-_22219738 | 0.66 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr13_-_23755374 | 0.64 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr2_+_28082943 | 0.62 |
ENSMUST00000113920.8
|
Olfm1
|
olfactomedin 1 |
| chr13_+_38009981 | 0.62 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr13_-_54835996 | 0.61 |
ENSMUST00000150806.8
ENSMUST00000125927.8 |
Rnf44
|
ring finger protein 44 |
| chr2_-_18053158 | 0.58 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chr3_-_58792633 | 0.57 |
ENSMUST00000055636.13
ENSMUST00000072551.7 ENSMUST00000051408.8 |
Clrn1
|
clarin 1 |
| chr7_-_15781838 | 0.57 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr13_-_54835878 | 0.55 |
ENSMUST00000125871.8
|
Rnf44
|
ring finger protein 44 |
| chr2_+_71884943 | 0.53 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_+_71127540 | 0.53 |
ENSMUST00000022699.10
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2 |
| chr16_-_92494203 | 0.53 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr2_+_28083105 | 0.52 |
ENSMUST00000100244.10
|
Olfm1
|
olfactomedin 1 |
| chr13_-_95661726 | 0.51 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr17_-_35383867 | 0.51 |
ENSMUST00000025253.12
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr2_-_18053595 | 0.51 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr13_-_22017677 | 0.51 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr2_-_45000389 | 0.50 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr18_+_69726654 | 0.50 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr6_+_136931519 | 0.49 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr3_+_45332831 | 0.47 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr12_+_71095112 | 0.47 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_79675727 | 0.45 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr13_+_41040657 | 0.44 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr8_-_85500010 | 0.44 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr13_-_22227114 | 0.44 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr1_+_143516402 | 0.43 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr19_+_13594739 | 0.43 |
ENSMUST00000217061.3
ENSMUST00000209005.4 ENSMUST00000208347.3 |
Olfr1487
|
olfactory receptor 1487 |
| chr7_-_78228116 | 0.42 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr19_+_13608985 | 0.42 |
ENSMUST00000217182.3
|
Olfr1489
|
olfactory receptor 1489 |
| chr11_+_115272732 | 0.41 |
ENSMUST00000053288.6
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr12_-_114252202 | 0.41 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chrX_-_16777913 | 0.40 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr10_+_7708178 | 0.40 |
ENSMUST00000039484.6
|
Zc3h12d
|
zinc finger CCCH type containing 12D |
| chr13_+_41071077 | 0.40 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr5_+_75316552 | 0.39 |
ENSMUST00000168162.5
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr6_+_136495784 | 0.39 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_-_59778560 | 0.39 |
ENSMUST00000153136.2
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chrX_+_7744535 | 0.39 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr1_-_87322443 | 0.38 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr13_+_23758555 | 0.38 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr10_-_12490424 | 0.38 |
ENSMUST00000217994.2
|
Utrn
|
utrophin |
| chr19_-_40982576 | 0.37 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr13_+_5911481 | 0.37 |
ENSMUST00000000080.8
|
Klf6
|
Kruppel-like factor 6 |
| chr13_-_54836077 | 0.37 |
ENSMUST00000150626.2
ENSMUST00000134177.8 |
Rnf44
|
ring finger protein 44 |
| chr11_+_44508137 | 0.37 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr1_-_143879738 | 0.37 |
ENSMUST00000153527.3
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr13_-_54835508 | 0.36 |
ENSMUST00000177950.8
ENSMUST00000146931.8 |
Rnf44
|
ring finger protein 44 |
| chrX_+_80114242 | 0.36 |
ENSMUST00000171953.8
ENSMUST00000026760.3 |
Tmem47
|
transmembrane protein 47 |
| chr9_+_86625694 | 0.35 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr4_+_97660971 | 0.35 |
ENSMUST00000152023.8
|
Nfia
|
nuclear factor I/A |
| chr11_-_98666159 | 0.34 |
ENSMUST00000064941.7
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr4_-_128699838 | 0.33 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr11_-_115968745 | 0.33 |
ENSMUST00000156545.2
ENSMUST00000075036.9 ENSMUST00000106451.8 |
Unc13d
|
unc-13 homolog D |
| chr4_-_24800890 | 0.33 |
ENSMUST00000108214.9
|
Klhl32
|
kelch-like 32 |
| chr6_+_68402550 | 0.33 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr5_+_42225303 | 0.31 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr7_-_45546081 | 0.31 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr7_+_110372860 | 0.31 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chrX_-_139443926 | 0.31 |
ENSMUST00000055738.12
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr7_+_79676095 | 0.31 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chr11_+_74721733 | 0.30 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr18_+_69726431 | 0.30 |
ENSMUST00000201091.4
ENSMUST00000201037.4 ENSMUST00000114977.5 |
Tcf4
|
transcription factor 4 |
| chr1_+_128069677 | 0.29 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr5_+_21748523 | 0.29 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr4_+_105014536 | 0.28 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr13_+_83672965 | 0.28 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr5_+_104447037 | 0.27 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr4_-_62278761 | 0.27 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr10_+_79763164 | 0.27 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr8_+_71207326 | 0.27 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr6_-_70412460 | 0.27 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr7_+_44667377 | 0.26 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr7_+_24981604 | 0.26 |
ENSMUST00000163320.8
ENSMUST00000005578.13 |
Cic
|
capicua transcriptional repressor |
| chr5_-_142880987 | 0.26 |
ENSMUST00000035985.8
|
Fbxl18
|
F-box and leucine-rich repeat protein 18 |
| chr17_+_47451868 | 0.26 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr16_+_91526169 | 0.25 |
ENSMUST00000114001.8
ENSMUST00000113999.8 ENSMUST00000064797.12 ENSMUST00000114002.9 ENSMUST00000095909.10 ENSMUST00000056482.14 ENSMUST00000113996.8 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr13_+_102830029 | 0.24 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr12_-_56583582 | 0.24 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr2_-_160950936 | 0.24 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr6_+_68247469 | 0.24 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr6_+_68026941 | 0.23 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr9_-_102231884 | 0.22 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr6_+_92069376 | 0.22 |
ENSMUST00000113463.8
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr8_+_23464860 | 0.22 |
ENSMUST00000110688.9
ENSMUST00000121802.9 |
Ank1
|
ankyrin 1, erythroid |
| chr5_+_138083345 | 0.22 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr8_+_66838927 | 0.22 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr2_-_45001141 | 0.22 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_144525255 | 0.22 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr2_+_90927053 | 0.22 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr12_-_114752425 | 0.21 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr8_+_121215155 | 0.21 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr2_+_121120070 | 0.21 |
ENSMUST00000094639.10
|
Map1a
|
microtubule-associated protein 1 A |
| chr10_-_127456791 | 0.20 |
ENSMUST00000118455.2
ENSMUST00000121829.8 |
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr8_+_23548541 | 0.20 |
ENSMUST00000173248.8
|
Ank1
|
ankyrin 1, erythroid |
| chr4_-_152561896 | 0.20 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr2_-_32977182 | 0.20 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr2_-_111908072 | 0.20 |
ENSMUST00000213577.2
ENSMUST00000216071.2 |
Olfr1313
|
olfactory receptor 1313 |
| chr18_+_69726055 | 0.20 |
ENSMUST00000114978.9
|
Tcf4
|
transcription factor 4 |
| chr1_-_64160557 | 0.20 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr2_-_45007407 | 0.20 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_70292451 | 0.19 |
ENSMUST00000103387.3
|
Igkv8-21
|
immunoglobulin kappa variable 8-21 |
| chr1_-_163141230 | 0.18 |
ENSMUST00000174397.3
ENSMUST00000075805.13 |
Prrx1
|
paired related homeobox 1 |
| chr17_+_41121979 | 0.18 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr6_+_68233361 | 0.18 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr5_+_76988444 | 0.18 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr10_-_8761777 | 0.18 |
ENSMUST00000015449.6
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr17_-_24752683 | 0.18 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr17_-_36220518 | 0.18 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr6_+_70675416 | 0.18 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr10_+_79762858 | 0.18 |
ENSMUST00000019708.12
ENSMUST00000105377.8 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr6_+_122684448 | 0.18 |
ENSMUST00000112581.8
ENSMUST00000112580.8 ENSMUST00000012540.5 |
Nanog
|
Nanog homeobox |
| chr7_-_28071919 | 0.18 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr3_-_84128160 | 0.17 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr4_-_62278673 | 0.17 |
ENSMUST00000084527.10
ENSMUST00000098033.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr6_+_68279392 | 0.17 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr6_+_108190163 | 0.17 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr15_+_12824925 | 0.17 |
ENSMUST00000090292.13
|
Drosha
|
drosha, ribonuclease type III |
| chr1_-_66856695 | 0.17 |
ENSMUST00000068168.10
ENSMUST00000113987.2 |
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr2_+_51928017 | 0.17 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr5_+_105879914 | 0.17 |
ENSMUST00000154807.2
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr12_-_115876396 | 0.17 |
ENSMUST00000103547.2
|
Ighv1-80
|
immunoglobulin heavy variable 1-80 |
| chr2_-_26393826 | 0.16 |
ENSMUST00000028288.5
|
Notch1
|
notch 1 |
| chr9_-_103980200 | 0.16 |
ENSMUST00000188000.2
|
Ackr4
|
atypical chemokine receptor 4 |
| chr14_-_103220066 | 0.16 |
ENSMUST00000184744.2
|
Kctd12
|
potassium channel tetramerisation domain containing 12 |
| chr12_-_113552322 | 0.16 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr2_-_25982160 | 0.16 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr7_-_44702269 | 0.15 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chr13_-_108026590 | 0.15 |
ENSMUST00000225822.4
ENSMUST00000225197.3 |
Zswim6
|
zinc finger SWIM-type containing 6 |
| chr8_-_34237752 | 0.15 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr6_-_70383976 | 0.15 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr6_-_70121150 | 0.15 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chrX_+_165127688 | 0.15 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr13_+_14238361 | 0.15 |
ENSMUST00000129488.8
ENSMUST00000110536.8 ENSMUST00000110534.8 ENSMUST00000039538.15 ENSMUST00000110533.2 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr6_+_108190050 | 0.15 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chrX_+_93278203 | 0.14 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15 |
| chr11_-_48708159 | 0.14 |
ENSMUST00000047145.14
|
Trim41
|
tripartite motif-containing 41 |
| chr14_+_20398230 | 0.14 |
ENSMUST00000224930.2
ENSMUST00000224110.2 ENSMUST00000225942.2 ENSMUST00000051915.7 ENSMUST00000090499.13 ENSMUST00000224721.2 ENSMUST00000090503.12 ENSMUST00000225991.2 ENSMUST00000037698.13 |
Fam149b
|
family with sequence similarity 149, member B |
| chr12_+_52746158 | 0.14 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr7_+_63835285 | 0.14 |
ENSMUST00000206263.2
ENSMUST00000206107.2 ENSMUST00000205731.2 ENSMUST00000206706.2 ENSMUST00000205690.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr16_-_16962279 | 0.14 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr2_-_35995283 | 0.14 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr5_+_111565129 | 0.13 |
ENSMUST00000094463.5
|
Mn1
|
meningioma 1 |
| chr6_+_70648743 | 0.13 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr7_+_110373447 | 0.13 |
ENSMUST00000147587.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr6_+_68414401 | 0.13 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr7_-_28072022 | 0.13 |
ENSMUST00000144700.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr10_+_128542120 | 0.13 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chr1_-_91386976 | 0.13 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr1_-_163141278 | 0.12 |
ENSMUST00000027878.14
|
Prrx1
|
paired related homeobox 1 |
| chr2_+_30485048 | 0.12 |
ENSMUST00000102853.4
|
Cstad
|
CSA-conditional, T cell activation-dependent protein |
| chr6_-_55658242 | 0.12 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr11_-_3454766 | 0.12 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr16_+_10306075 | 0.12 |
ENSMUST00000023147.8
|
Ciita
|
class II transactivator |
| chr4_+_136150835 | 0.12 |
ENSMUST00000088677.5
ENSMUST00000121571.8 ENSMUST00000117699.2 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr12_-_113625906 | 0.12 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr12_-_114878652 | 0.12 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr13_+_102830104 | 0.12 |
ENSMUST00000172138.2
|
Cd180
|
CD180 antigen |
| chr2_+_57128309 | 0.11 |
ENSMUST00000112618.9
ENSMUST00000028167.3 |
Gpd2
|
glycerol phosphate dehydrogenase 2, mitochondrial |
| chr8_+_71069476 | 0.11 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr15_+_57558048 | 0.11 |
ENSMUST00000096430.11
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr14_-_39194782 | 0.11 |
ENSMUST00000168810.9
ENSMUST00000173780.2 ENSMUST00000166968.9 |
Nrg3
|
neuregulin 3 |
| chr9_+_123901979 | 0.11 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr7_+_18962301 | 0.11 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr4_-_63965161 | 0.11 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr1_-_143879877 | 0.11 |
ENSMUST00000127206.8
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr12_-_114443071 | 0.11 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou5f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.2 | 0.5 | GO:1900135 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.1 | 0.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.5 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.9 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.4 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.4 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 1.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.4 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.1 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 0.1 | 0.8 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.5 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.1 | 0.3 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.6 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.3 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.2 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.0 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.7 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:0050882 | epithelial fluid transport(GO:0042045) voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.7 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.5 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.8 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 1.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.3 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 1.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |