Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Pou6f2_Pou4f2
Z-value: 1.25
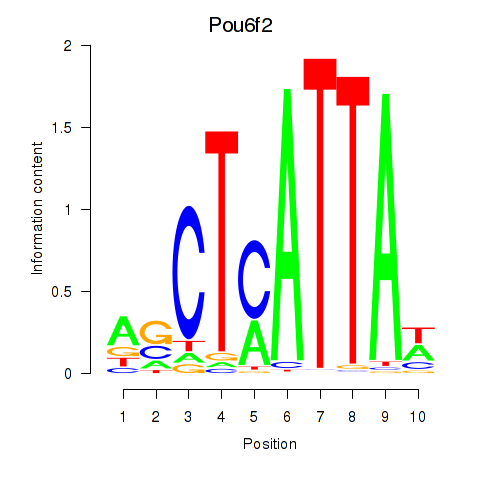
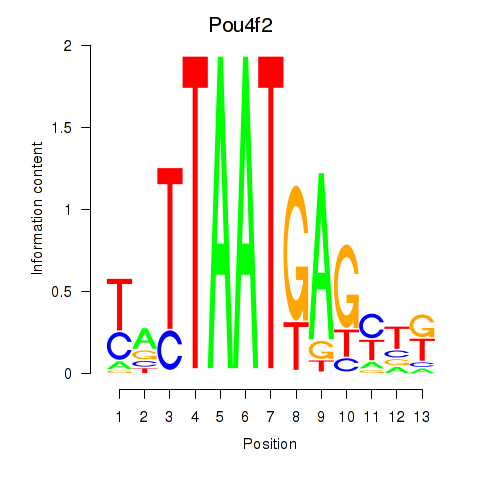
Transcription factors associated with Pou6f2_Pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou6f2
|
ENSMUSG00000009734.19 | POU domain, class 6, transcription factor 2 |
|
Pou4f2
|
ENSMUSG00000031688.5 | POU domain, class 4, transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou6f2 | mm39_v1_chr13_-_18556626_18556626 | 0.90 | 3.7e-02 | Click! |
| Pou4f2 | mm39_v1_chr8_-_79163269_79163278 | 0.56 | 3.2e-01 | Click! |
Activity profile of Pou6f2_Pou4f2 motif
Sorted Z-values of Pou6f2_Pou4f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_111820618 | 1.48 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr12_-_99529767 | 1.06 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr2_-_111843053 | 1.03 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chrX_+_16485937 | 0.99 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr14_-_50586329 | 0.94 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr2_+_37101586 | 0.90 |
ENSMUST00000214897.2
|
Olfr366
|
olfactory receptor 366 |
| chr7_-_6525801 | 0.77 |
ENSMUST00000213504.2
ENSMUST00000216447.2 ENSMUST00000213656.2 ENSMUST00000207820.3 |
Olfr1349
|
olfactory receptor 1349 |
| chr11_+_73851643 | 0.73 |
ENSMUST00000213134.2
ENSMUST00000216291.2 |
Olfr397
|
olfactory receptor 397 |
| chr13_-_56444118 | 0.73 |
ENSMUST00000224801.2
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr5_-_62923463 | 0.72 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr8_-_5155347 | 0.60 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr17_-_37399343 | 0.59 |
ENSMUST00000207101.2
ENSMUST00000217397.2 ENSMUST00000215195.2 ENSMUST00000216488.2 |
Olfr90
|
olfactory receptor 90 |
| chr14_-_51134930 | 0.56 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr7_+_23239157 | 0.53 |
ENSMUST00000235361.2
|
Vmn1r168
|
vomeronasal 1 receptor 168 |
| chr10_+_4284536 | 0.46 |
ENSMUST00000239373.2
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr2_+_88644840 | 0.44 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr7_+_23752679 | 0.43 |
ENSMUST00000236959.2
|
Vmn1r183
|
vomeronasal 1 receptor 183 |
| chr6_-_41681273 | 0.43 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr15_+_35371644 | 0.41 |
ENSMUST00000227455.2
|
Vps13b
|
vacuolar protein sorting 13B |
| chr7_+_23400128 | 0.39 |
ENSMUST00000226233.2
ENSMUST00000227987.2 |
Vmn1r173
|
vomeronasal 1 receptor 173 |
| chr7_-_106531426 | 0.38 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr19_+_13890894 | 0.35 |
ENSMUST00000216623.2
ENSMUST00000216835.2 |
Olfr1505
|
olfactory receptor 1505 |
| chr7_+_23451695 | 0.34 |
ENSMUST00000228331.2
|
Vmn1r174
|
vomeronasal 1 receptor 174 |
| chr14_+_26761023 | 0.33 |
ENSMUST00000223942.2
|
Il17rd
|
interleukin 17 receptor D |
| chr2_-_58050494 | 0.31 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr2_+_96148418 | 0.28 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr13_-_19917092 | 0.28 |
ENSMUST00000151029.3
|
Gpr141b
|
G protein-coupled receptor 141B |
| chr2_+_172994841 | 0.27 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr2_-_88157559 | 0.24 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr12_-_56581823 | 0.24 |
ENSMUST00000178477.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr3_-_88363704 | 0.23 |
ENSMUST00000141471.2
ENSMUST00000123753.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr2_-_153079828 | 0.22 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr16_+_43067641 | 0.22 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_172168764 | 0.22 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr8_+_4288815 | 0.22 |
ENSMUST00000003027.14
ENSMUST00000110999.8 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr7_-_10292412 | 0.21 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr7_+_126184108 | 0.21 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr16_+_43323970 | 0.21 |
ENSMUST00000126100.8
ENSMUST00000123047.8 ENSMUST00000156981.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr18_-_38068430 | 0.19 |
ENSMUST00000025337.14
|
Diaph1
|
diaphanous related formin 1 |
| chr3_+_84500854 | 0.19 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr15_-_50746202 | 0.19 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr18_-_15197138 | 0.19 |
ENSMUST00000234864.2
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr19_+_42034231 | 0.18 |
ENSMUST00000172244.8
ENSMUST00000081714.5 |
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr7_-_102566717 | 0.17 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr1_-_192880260 | 0.16 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr8_+_84262409 | 0.16 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr8_+_4288733 | 0.16 |
ENSMUST00000110998.9
ENSMUST00000062686.11 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr18_-_38068456 | 0.16 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr13_-_21744063 | 0.16 |
ENSMUST00000217519.2
|
Olfr1535
|
olfactory receptor 1535 |
| chr19_+_55730696 | 0.16 |
ENSMUST00000153888.9
ENSMUST00000127233.9 ENSMUST00000061496.17 ENSMUST00000111656.8 ENSMUST00000111657.11 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr1_-_143652711 | 0.15 |
ENSMUST00000159879.2
|
Ro60
|
Ro60, Y RNA binding protein |
| chr6_+_125529911 | 0.15 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr18_+_20509771 | 0.15 |
ENSMUST00000076737.7
|
Dsg1b
|
desmoglein 1 beta |
| chr11_+_73241609 | 0.14 |
ENSMUST00000120137.3
|
Olfr20
|
olfactory receptor 20 |
| chr14_-_20718337 | 0.14 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chrX_-_58211440 | 0.13 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr6_-_146855880 | 0.13 |
ENSMUST00000111622.2
ENSMUST00000036592.15 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr2_-_73283010 | 0.12 |
ENSMUST00000151939.2
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr12_+_52746158 | 0.11 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr10_+_127256736 | 0.11 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr4_-_133329479 | 0.11 |
ENSMUST00000057311.4
|
Sfn
|
stratifin |
| chr2_+_84564394 | 0.11 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr6_+_134958681 | 0.11 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr4_-_43710231 | 0.10 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr6_-_41752111 | 0.10 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr14_+_26760898 | 0.10 |
ENSMUST00000035336.5
|
Il17rd
|
interleukin 17 receptor D |
| chr10_+_40225272 | 0.10 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr3_-_37286714 | 0.10 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr1_+_153541020 | 0.09 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr5_+_117979899 | 0.09 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr17_-_49871291 | 0.09 |
ENSMUST00000224595.2
ENSMUST00000057610.8 |
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr5_+_135135735 | 0.09 |
ENSMUST00000201977.4
ENSMUST00000005507.10 |
Mlxipl
|
MLX interacting protein-like |
| chr10_-_95158827 | 0.09 |
ENSMUST00000220279.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr17_+_34811217 | 0.09 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr4_+_117706390 | 0.08 |
ENSMUST00000132043.9
ENSMUST00000169990.8 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr16_+_56024676 | 0.08 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr8_+_84393263 | 0.08 |
ENSMUST00000019608.7
|
Ptger1
|
prostaglandin E receptor 1 (subtype EP1) |
| chr7_-_86016045 | 0.08 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr11_+_67090878 | 0.08 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr7_-_73191484 | 0.07 |
ENSMUST00000197642.2
ENSMUST00000026895.14 ENSMUST00000169922.9 |
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr14_-_75991903 | 0.07 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr7_-_126183716 | 0.07 |
ENSMUST00000150311.8
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr10_+_127256993 | 0.07 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr9_-_19289965 | 0.07 |
ENSMUST00000216839.3
|
Olfr847
|
olfactory receptor 847 |
| chr4_+_117706559 | 0.07 |
ENSMUST00000163288.2
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_115630282 | 0.06 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chrX_-_74460137 | 0.06 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr2_-_79287095 | 0.06 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr7_-_28038129 | 0.06 |
ENSMUST00000209141.2
ENSMUST00000003527.10 |
Supt5
|
suppressor of Ty 5, DSIF elongation factor subunit |
| chr16_-_45544960 | 0.06 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr9_-_99302205 | 0.06 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr7_+_28580847 | 0.06 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr17_+_17622934 | 0.06 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr8_-_3674993 | 0.05 |
ENSMUST00000142431.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chrX_+_82898974 | 0.04 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chrX_-_138683102 | 0.04 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr19_+_55730242 | 0.04 |
ENSMUST00000111662.11
ENSMUST00000041717.14 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr12_-_81579614 | 0.03 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr19_-_5610628 | 0.03 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr1_+_153541412 | 0.03 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr14_+_63673843 | 0.03 |
ENSMUST00000121288.2
|
Fam167a
|
family with sequence similarity 167, member A |
| chr17_+_85335775 | 0.03 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr5_-_90788323 | 0.02 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_-_79645101 | 0.02 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr2_+_3115250 | 0.02 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr17_-_31417932 | 0.02 |
ENSMUST00000114549.4
|
Tmprss3
|
transmembrane protease, serine 3 |
| chr14_-_51134906 | 0.02 |
ENSMUST00000170855.2
|
Klhl33
|
kelch-like 33 |
| chr2_-_92222979 | 0.01 |
ENSMUST00000111279.9
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr19_+_55730488 | 0.01 |
ENSMUST00000111659.9
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chrX_+_99019176 | 0.01 |
ENSMUST00000113781.8
ENSMUST00000113783.8 ENSMUST00000113779.8 ENSMUST00000113776.8 ENSMUST00000113775.8 ENSMUST00000113780.8 ENSMUST00000113778.8 ENSMUST00000113777.8 ENSMUST00000071453.3 |
Eda
|
ectodysplasin-A |
| chr7_-_126183392 | 0.01 |
ENSMUST00000128970.8
ENSMUST00000116269.9 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr18_-_55123153 | 0.01 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608 |
| chr17_-_3608056 | 0.01 |
ENSMUST00000041003.8
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr7_+_143729250 | 0.01 |
ENSMUST00000105900.9
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr4_+_102446883 | 0.01 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr5_+_29400981 | 0.01 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr11_+_61376257 | 0.01 |
ENSMUST00000064783.10
ENSMUST00000040522.7 |
Mfap4
|
microfibrillar-associated protein 4 |
| chr4_+_34893772 | 0.01 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr13_-_103901010 | 0.01 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr10_+_101517348 | 0.01 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr17_+_6026015 | 0.00 |
ENSMUST00000115790.8
|
Synj2
|
synaptojanin 2 |
| chr4_+_118577335 | 0.00 |
ENSMUST00000217334.3
ENSMUST00000213436.3 ENSMUST00000216242.3 |
Olfr1340
|
olfactory receptor 1340 |
| chr19_+_55730316 | 0.00 |
ENSMUST00000111658.10
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr8_-_3675024 | 0.00 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr11_+_115725140 | 0.00 |
ENSMUST00000173289.8
ENSMUST00000137900.2 |
Llgl2
|
LLGL2 scribble cell polarity complex component |
| chr8_-_3675274 | 0.00 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr15_+_102845192 | 0.00 |
ENSMUST00000055562.3
|
Hoxc12
|
homeobox C12 |
| chr10_+_102348076 | 0.00 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr19_-_18978463 | 0.00 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou6f2_Pou4f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.4 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.3 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.1 | 0.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 1.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.2 | GO:0048625 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.0 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.1 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 4.7 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.3 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |