Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Ppara
Z-value: 0.94
Transcription factors associated with Ppara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppara
|
ENSMUSG00000022383.14 | peroxisome proliferator activated receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppara | mm39_v1_chr15_+_85619765_85619817 | 0.69 | 2.0e-01 | Click! |
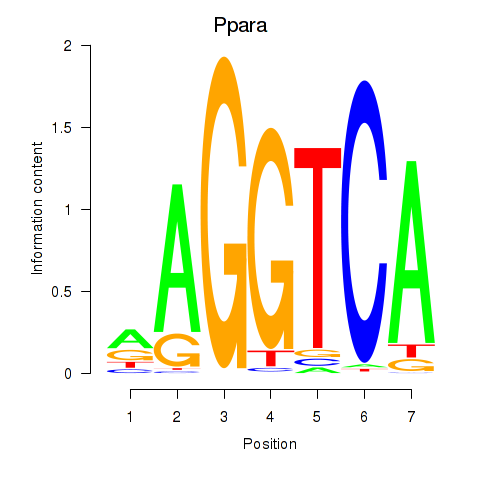
Activity profile of Ppara motif
Sorted Z-values of Ppara motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_129854256 | 1.05 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr1_-_140111138 | 0.98 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 0.89 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr13_-_12355604 | 0.67 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr6_+_79794899 | 0.67 |
ENSMUST00000179797.3
|
Gm20594
|
predicted gene, 20594 |
| chr5_+_52521133 | 0.66 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr11_-_96868483 | 0.41 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr15_-_5273659 | 0.38 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr6_-_52194414 | 0.37 |
ENSMUST00000140316.2
|
Hoxa7
|
homeobox A7 |
| chr15_-_5273645 | 0.37 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr5_-_66309244 | 0.36 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr5_-_23881353 | 0.36 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr2_+_71811526 | 0.35 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrX_-_133062677 | 0.34 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr8_-_70975734 | 0.33 |
ENSMUST00000137610.3
|
Kxd1
|
KxDL motif containing 1 |
| chr15_-_42540363 | 0.33 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr6_-_99005835 | 0.33 |
ENSMUST00000154163.9
|
Foxp1
|
forkhead box P1 |
| chr5_-_66308421 | 0.32 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr17_-_36220518 | 0.32 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chrM_-_14061 | 0.32 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrM_+_14138 | 0.32 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr5_+_16758538 | 0.31 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chr8_-_70975813 | 0.31 |
ENSMUST00000121623.8
ENSMUST00000093456.12 ENSMUST00000118850.8 |
Kxd1
|
KxDL motif containing 1 |
| chr6_-_142453531 | 0.31 |
ENSMUST00000134191.3
ENSMUST00000239397.2 ENSMUST00000239395.2 ENSMUST00000032373.12 |
Ldhb
|
lactate dehydrogenase B |
| chr4_-_3973586 | 0.31 |
ENSMUST00000089430.6
|
Gm11808
|
predicted gene 11808 |
| chr16_-_18066591 | 0.31 |
ENSMUST00000115645.10
|
Ranbp1
|
RAN binding protein 1 |
| chrX_+_72760183 | 0.30 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr5_+_16758777 | 0.30 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr7_-_19410749 | 0.29 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr12_-_111638722 | 0.29 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr4_-_131802606 | 0.27 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr5_+_16758897 | 0.27 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr6_-_99005212 | 0.26 |
ENSMUST00000177437.8
ENSMUST00000177229.8 ENSMUST00000124058.8 |
Foxp1
|
forkhead box P1 |
| chr6_-_52194440 | 0.26 |
ENSMUST00000048715.9
|
Hoxa7
|
homeobox A7 |
| chr10_+_57670431 | 0.26 |
ENSMUST00000151623.8
ENSMUST00000020022.8 |
Smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr11_+_116324913 | 0.26 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr5_-_66308666 | 0.25 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr1_+_74324089 | 0.25 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr15_-_35938328 | 0.25 |
ENSMUST00000014457.15
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr3_-_129834788 | 0.25 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr7_+_79992839 | 0.24 |
ENSMUST00000032747.7
ENSMUST00000206480.2 ENSMUST00000206074.2 ENSMUST00000206122.2 |
Hddc3
|
HD domain containing 3 |
| chr19_-_3962733 | 0.24 |
ENSMUST00000075092.8
ENSMUST00000235847.2 ENSMUST00000235301.2 ENSMUST00000237341.2 |
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr15_-_77653979 | 0.24 |
ENSMUST00000229259.2
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr11_+_98754434 | 0.24 |
ENSMUST00000142414.8
ENSMUST00000037480.9 |
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr9_+_39932760 | 0.24 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr14_+_20732804 | 0.24 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr13_-_93328619 | 0.24 |
ENSMUST00000224464.2
|
Tent2
|
terminal nucleotidyltransferase 2 |
| chr12_+_33364288 | 0.24 |
ENSMUST00000144586.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr15_-_66432938 | 0.24 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr15_-_79688910 | 0.23 |
ENSMUST00000175858.10
ENSMUST00000023057.10 |
Nptxr
|
neuronal pentraxin receptor |
| chr7_+_29931309 | 0.23 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr1_-_73967664 | 0.23 |
ENSMUST00000187691.7
|
Tns1
|
tensin 1 |
| chrM_+_3906 | 0.23 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr15_-_74624811 | 0.23 |
ENSMUST00000189128.2
ENSMUST00000023259.15 |
Lynx1
|
Ly6/neurotoxin 1 |
| chr1_+_6284823 | 0.23 |
ENSMUST00000027040.13
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr1_-_167221344 | 0.22 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr11_-_54924231 | 0.22 |
ENSMUST00000108883.10
ENSMUST00000102727.3 |
Anxa6
|
annexin A6 |
| chr17_+_15163466 | 0.22 |
ENSMUST00000164837.3
ENSMUST00000174004.2 |
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene |
| chr14_+_58308004 | 0.21 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr16_-_89940652 | 0.21 |
ENSMUST00000114124.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr2_-_5680801 | 0.21 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr5_+_34527230 | 0.21 |
ENSMUST00000180376.8
|
Fam193a
|
family with sequence homology 193, member A |
| chr19_-_7194912 | 0.21 |
ENSMUST00000039758.6
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr10_-_67972401 | 0.21 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr16_+_10363222 | 0.21 |
ENSMUST00000155633.8
ENSMUST00000066345.15 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr1_+_131838220 | 0.21 |
ENSMUST00000189946.7
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr10_-_93425553 | 0.21 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr9_-_43027809 | 0.21 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr7_+_110368037 | 0.20 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr6_-_52181393 | 0.20 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr8_+_13455080 | 0.20 |
ENSMUST00000033827.8
ENSMUST00000209909.2 |
Grk1
|
G protein-coupled receptor kinase 1 |
| chr14_+_25459690 | 0.20 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr12_-_111933328 | 0.20 |
ENSMUST00000222874.2
ENSMUST00000223191.2 ENSMUST00000021719.7 |
Atp5mpl
|
ATP synthase membrane subunit 6.8PL |
| chr2_+_145627900 | 0.20 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr13_-_86194889 | 0.20 |
ENSMUST00000131011.2
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr1_+_32211792 | 0.20 |
ENSMUST00000027226.12
ENSMUST00000189878.2 ENSMUST00000188257.7 ENSMUST00000185666.2 |
Khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr19_-_6957789 | 0.19 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr11_+_75400889 | 0.19 |
ENSMUST00000042972.7
|
Rilp
|
Rab interacting lysosomal protein |
| chr8_-_84321032 | 0.19 |
ENSMUST00000163837.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr17_-_36220924 | 0.19 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr17_-_35407403 | 0.19 |
ENSMUST00000097336.5
|
Lst1
|
leukocyte specific transcript 1 |
| chr6_-_125143425 | 0.19 |
ENSMUST00000117757.9
ENSMUST00000073605.15 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr7_-_80052491 | 0.19 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr17_-_45125468 | 0.19 |
ENSMUST00000159943.8
ENSMUST00000160673.8 |
Runx2
|
runt related transcription factor 2 |
| chr19_-_47079052 | 0.18 |
ENSMUST00000235771.2
ENSMUST00000096014.5 ENSMUST00000236170.2 |
Atp5md
|
ATP synthase membrane subunit DAPIT |
| chr7_+_3620356 | 0.18 |
ENSMUST00000076657.11
ENSMUST00000108644.8 |
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr9_+_50515207 | 0.18 |
ENSMUST00000044051.6
|
Timm8b
|
translocase of inner mitochondrial membrane 8B |
| chr1_-_170886924 | 0.18 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr7_+_27879650 | 0.18 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr18_+_60936910 | 0.18 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr5_+_31855009 | 0.18 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr10_+_93983844 | 0.18 |
ENSMUST00000105290.9
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr5_-_108582239 | 0.18 |
ENSMUST00000049628.16
ENSMUST00000118632.2 |
Atp5k
|
ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E |
| chr19_-_5713701 | 0.17 |
ENSMUST00000164304.9
ENSMUST00000237544.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr7_+_3339059 | 0.17 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr4_+_140688514 | 0.17 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_-_63970270 | 0.17 |
ENSMUST00000049091.9
|
Cox10
|
heme A:farnesyltransferase cytochrome c oxidase assembly factor 10 |
| chr8_+_95703728 | 0.17 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr2_-_45007407 | 0.17 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_172327569 | 0.17 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr8_+_71921824 | 0.17 |
ENSMUST00000124745.8
ENSMUST00000138892.2 ENSMUST00000147642.2 |
Dda1
|
DET1 and DDB1 associated 1 |
| chr11_-_74480870 | 0.17 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr10_-_80895949 | 0.17 |
ENSMUST00000219401.2
ENSMUST00000220317.2 ENSMUST00000005067.6 ENSMUST00000218208.2 |
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr2_+_85545763 | 0.17 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr2_+_71884943 | 0.17 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_28834391 | 0.16 |
ENSMUST00000160194.8
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr16_+_10363203 | 0.16 |
ENSMUST00000115824.10
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr7_-_30259253 | 0.16 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr15_-_98118858 | 0.16 |
ENSMUST00000142443.8
ENSMUST00000170618.8 |
Gm44579
Olfr287
|
predicted gene 44579 olfactory receptor 287 |
| chr11_-_3454766 | 0.16 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr7_+_3339077 | 0.16 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr1_-_183003077 | 0.16 |
ENSMUST00000194033.2
|
Disp1
|
dispatched RND transporter family member 1 |
| chr2_+_70491901 | 0.16 |
ENSMUST00000112201.8
ENSMUST00000028509.11 ENSMUST00000133432.8 ENSMUST00000112205.2 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr6_-_142910094 | 0.16 |
ENSMUST00000032421.4
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr10_+_80100812 | 0.16 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr17_+_48047955 | 0.16 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr2_+_61423421 | 0.16 |
ENSMUST00000112495.8
ENSMUST00000112501.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr16_-_18884431 | 0.16 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr6_+_22288220 | 0.16 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr15_+_84553801 | 0.16 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr9_-_110474398 | 0.16 |
ENSMUST00000149089.2
|
Nbeal2
|
neurobeachin-like 2 |
| chrX_-_12026594 | 0.16 |
ENSMUST00000043441.13
|
Bcor
|
BCL6 interacting corepressor |
| chr15_-_79281056 | 0.15 |
ENSMUST00000228472.2
|
Tmem184b
|
transmembrane protein 184b |
| chr11_-_106679671 | 0.15 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr16_+_84631789 | 0.15 |
ENSMUST00000114184.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr9_-_110818679 | 0.15 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr14_+_58035640 | 0.15 |
ENSMUST00000111269.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr15_-_63932288 | 0.15 |
ENSMUST00000063838.11
ENSMUST00000228908.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr11_-_53321606 | 0.15 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr16_-_43960045 | 0.15 |
ENSMUST00000147025.2
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr5_-_77243123 | 0.15 |
ENSMUST00000113453.9
|
Hopx
|
HOP homeobox |
| chr15_-_76113692 | 0.15 |
ENSMUST00000074834.12
|
Plec
|
plectin |
| chr7_-_24705320 | 0.15 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr11_-_4654303 | 0.14 |
ENSMUST00000058407.6
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr15_-_103242697 | 0.14 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr11_+_85243970 | 0.14 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr18_-_15196612 | 0.14 |
ENSMUST00000168989.9
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr5_-_106844396 | 0.14 |
ENSMUST00000137285.8
ENSMUST00000124263.2 ENSMUST00000112695.4 ENSMUST00000155495.8 ENSMUST00000135108.2 ENSMUST00000149128.3 |
Zfp644
Gm28039
|
zinc finger protein 644 predicted gene, 28039 |
| chr9_+_21914083 | 0.14 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr7_-_30259025 | 0.14 |
ENSMUST00000043975.11
ENSMUST00000156241.2 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr9_-_106769069 | 0.14 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr8_-_123405392 | 0.14 |
ENSMUST00000134045.2
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr17_-_24005020 | 0.14 |
ENSMUST00000024704.10
|
Flywch2
|
FLYWCH family member 2 |
| chr7_+_110367375 | 0.14 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr4_-_139695337 | 0.14 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr5_+_31855304 | 0.14 |
ENSMUST00000114515.9
|
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr2_-_7086018 | 0.14 |
ENSMUST00000114923.3
ENSMUST00000182706.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr8_-_70448872 | 0.14 |
ENSMUST00000177851.9
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr10_+_60185093 | 0.14 |
ENSMUST00000105459.2
|
Vsir
|
V-set immunoregulatory receptor |
| chr11_-_95966477 | 0.14 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr9_+_108337726 | 0.14 |
ENSMUST00000061209.7
ENSMUST00000193269.2 |
Ccdc71
|
coiled-coil domain containing 71 |
| chr1_-_23961379 | 0.14 |
ENSMUST00000027339.14
|
Smap1
|
small ArfGAP 1 |
| chr14_+_25694594 | 0.14 |
ENSMUST00000022419.7
|
Ppif
|
peptidylprolyl isomerase F (cyclophilin F) |
| chr13_+_108350923 | 0.13 |
ENSMUST00000022207.10
|
Elovl7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| chr13_-_18118736 | 0.13 |
ENSMUST00000009003.9
|
Rala
|
v-ral simian leukemia viral oncogene A (ras related) |
| chr6_-_39702127 | 0.13 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr17_+_87061117 | 0.13 |
ENSMUST00000024954.11
|
Epas1
|
endothelial PAS domain protein 1 |
| chr9_+_89791943 | 0.13 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr5_-_143846600 | 0.13 |
ENSMUST00000031613.11
ENSMUST00000100483.3 |
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr2_-_7400997 | 0.13 |
ENSMUST00000137733.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr8_-_70449018 | 0.13 |
ENSMUST00000065169.12
ENSMUST00000212478.2 |
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr14_+_25459630 | 0.13 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr10_+_84412490 | 0.13 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr15_+_79025523 | 0.13 |
ENSMUST00000040077.8
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr7_+_89281897 | 0.13 |
ENSMUST00000032856.13
|
Me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr8_+_95722289 | 0.13 |
ENSMUST00000211984.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr2_-_60711706 | 0.13 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr1_+_82702598 | 0.13 |
ENSMUST00000078332.13
ENSMUST00000073025.12 ENSMUST00000161648.8 ENSMUST00000160786.8 ENSMUST00000162003.8 |
Mff
|
mitochondrial fission factor |
| chr11_-_115698894 | 0.13 |
ENSMUST00000132780.8
|
Caskin2
|
CASK-interacting protein 2 |
| chr3_+_137573839 | 0.13 |
ENSMUST00000197711.2
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr17_+_34341766 | 0.13 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr5_-_106844685 | 0.13 |
ENSMUST00000127434.8
ENSMUST00000112696.8 ENSMUST00000112698.8 |
Zfp644
|
zinc finger protein 644 |
| chr2_+_25132941 | 0.13 |
ENSMUST00000114355.2
ENSMUST00000060818.2 |
Rnf208
|
ring finger protein 208 |
| chr8_-_105350898 | 0.13 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr2_+_157870399 | 0.13 |
ENSMUST00000103123.10
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr9_+_115738523 | 0.12 |
ENSMUST00000119291.8
ENSMUST00000069651.13 |
Gadl1
|
glutamate decarboxylase-like 1 |
| chr13_-_93328719 | 0.12 |
ENSMUST00000048702.7
|
Tent2
|
terminal nucleotidyltransferase 2 |
| chr8_+_95703506 | 0.12 |
ENSMUST00000212581.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr10_+_79984097 | 0.12 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr2_+_129435448 | 0.12 |
ENSMUST00000049262.14
ENSMUST00000163034.8 ENSMUST00000160276.2 |
Sirpa
|
signal-regulatory protein alpha |
| chr4_-_141327146 | 0.12 |
ENSMUST00000141518.8
ENSMUST00000127455.8 ENSMUST00000105784.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr3_-_52012462 | 0.12 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3 |
| chr13_+_44883270 | 0.12 |
ENSMUST00000172830.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr6_+_123099615 | 0.12 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr10_+_94034817 | 0.12 |
ENSMUST00000020209.16
ENSMUST00000179990.8 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr11_+_74721733 | 0.12 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr11_-_99276868 | 0.12 |
ENSMUST00000211768.2
|
Krt10
|
keratin 10 |
| chr16_+_24540599 | 0.12 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_160714904 | 0.12 |
ENSMUST00000109460.8
ENSMUST00000127201.2 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr10_-_80691009 | 0.12 |
ENSMUST00000220225.2
ENSMUST00000035775.9 |
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_+_146156220 | 0.12 |
ENSMUST00000061937.13
ENSMUST00000029840.4 |
Ctbs
|
chitobiase |
| chr5_+_129802127 | 0.11 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr13_-_73476561 | 0.11 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr12_-_69728572 | 0.11 |
ENSMUST00000183277.8
ENSMUST00000035773.14 |
Sos2
|
SOS Ras/Rho guanine nucleotide exchange factor 2 |
| chr6_+_136509922 | 0.11 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr14_+_75373766 | 0.11 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr2_+_61423469 | 0.11 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr16_+_97157934 | 0.11 |
ENSMUST00000047275.8
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr10_-_43416941 | 0.11 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr3_-_95725944 | 0.11 |
ENSMUST00000200164.5
ENSMUST00000090791.8 ENSMUST00000197449.2 |
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppara
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:2000417 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.2 | 0.7 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.2 | 1.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.5 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.4 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.1 | 0.5 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.2 | GO:0061723 | glycophagy(GO:0061723) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.9 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:0060435 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.6 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 | 0.2 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.2 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.2 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.0 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.6 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.2 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 1.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 | 0.3 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.0 | 0.3 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.1 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.0 | 0.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.0 | 0.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |