Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
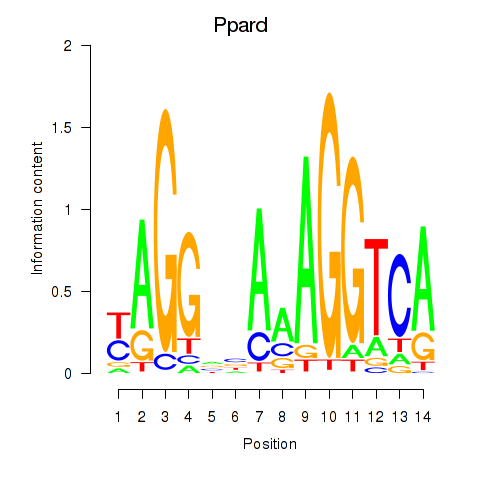
Results for Ppard
Z-value: 0.63
Transcription factors associated with Ppard
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppard
|
ENSMUSG00000002250.17 | peroxisome proliferator activator receptor delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppard | mm39_v1_chr17_+_28451674_28451764 | -0.55 | 3.4e-01 | Click! |
Activity profile of Ppard motif
Sorted Z-values of Ppard motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_56339915 | 0.70 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr11_-_69906171 | 0.37 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr3_+_51568588 | 0.36 |
ENSMUST00000099106.10
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr16_-_84632439 | 0.34 |
ENSMUST00000138279.2
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr7_+_43093507 | 0.32 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr2_-_73741664 | 0.31 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr19_-_6899173 | 0.30 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr15_+_101184488 | 0.30 |
ENSMUST00000229525.2
ENSMUST00000230525.2 |
Atg101
|
autophagy related 101 |
| chr8_-_58106057 | 0.28 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_-_32594156 | 0.28 |
ENSMUST00000127812.3
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr9_+_108539296 | 0.27 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr3_-_79535966 | 0.25 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr19_-_6899121 | 0.25 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr6_+_7844759 | 0.25 |
ENSMUST00000040159.6
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr3_+_51568625 | 0.25 |
ENSMUST00000159554.7
ENSMUST00000161590.4 |
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr4_+_118286898 | 0.24 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr1_-_63215812 | 0.24 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr1_-_120047868 | 0.23 |
ENSMUST00000112648.8
|
Dbi
|
diazepam binding inhibitor |
| chr19_-_32038838 | 0.22 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr3_-_105708601 | 0.22 |
ENSMUST00000197094.5
ENSMUST00000198004.2 |
Rap1a
|
RAS-related protein 1a |
| chr19_+_37184927 | 0.21 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr9_-_57169830 | 0.21 |
ENSMUST00000215298.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr4_+_118266582 | 0.21 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr2_+_92205651 | 0.21 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr3_-_83947416 | 0.20 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr1_+_171386752 | 0.20 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr5_+_36622342 | 0.20 |
ENSMUST00000031099.4
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr15_-_89310060 | 0.20 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr19_+_5510636 | 0.18 |
ENSMUST00000225141.2
ENSMUST00000025847.7 |
Fibp
|
fibroblast growth factor (acidic) intracellular binding protein |
| chr4_-_45108038 | 0.18 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr6_+_85408953 | 0.18 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr18_+_74912268 | 0.17 |
ENSMUST00000041053.11
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chr19_+_4644365 | 0.16 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr16_-_91728162 | 0.15 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr15_+_25774070 | 0.15 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr1_-_63215952 | 0.13 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr4_-_136776006 | 0.13 |
ENSMUST00000049583.8
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chr15_-_102097387 | 0.13 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr15_-_102097466 | 0.11 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr16_-_84632383 | 0.11 |
ENSMUST00000114191.8
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr3_-_79536166 | 0.10 |
ENSMUST00000029386.14
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr3_+_89958940 | 0.09 |
ENSMUST00000159064.8
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr15_-_27630724 | 0.09 |
ENSMUST00000059662.8
|
Otulin
|
OTU deubiquitinase with linear linkage specificity |
| chr2_-_164646794 | 0.09 |
ENSMUST00000103094.11
ENSMUST00000017451.7 |
Acot8
|
acyl-CoA thioesterase 8 |
| chr16_-_3725515 | 0.08 |
ENSMUST00000177221.2
ENSMUST00000177323.8 |
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr3_-_137837117 | 0.08 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr16_-_84632513 | 0.08 |
ENSMUST00000023608.14
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr3_-_8988854 | 0.08 |
ENSMUST00000042148.6
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr4_+_59581617 | 0.08 |
ENSMUST00000107528.8
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr2_+_84891281 | 0.06 |
ENSMUST00000238769.2
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr5_+_114284585 | 0.06 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr1_-_55127183 | 0.06 |
ENSMUST00000027123.15
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr1_-_133617824 | 0.06 |
ENSMUST00000189524.2
ENSMUST00000169295.8 |
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr19_+_10019023 | 0.06 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr1_-_55127312 | 0.06 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr9_-_45923908 | 0.06 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr5_-_31854942 | 0.05 |
ENSMUST00000031018.10
|
Rbks
|
ribokinase |
| chr1_+_180553569 | 0.05 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr11_+_4936824 | 0.05 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr15_-_89309998 | 0.04 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr10_-_125164399 | 0.04 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr17_-_34247016 | 0.04 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr1_+_74545203 | 0.04 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr19_+_4644425 | 0.04 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr2_+_158636727 | 0.04 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr18_-_36859732 | 0.04 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr1_+_63216281 | 0.03 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr17_-_27247581 | 0.03 |
ENSMUST00000143158.3
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr16_-_4442802 | 0.03 |
ENSMUST00000014445.7
|
Pam16
|
presequence translocase-asssociated motor 16 homolog (S. cerevisiae) |
| chr10_+_128139227 | 0.02 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr1_-_175806680 | 0.02 |
ENSMUST00000104983.2
|
Rbm8a2
|
RNA binding motif protein 8A2 |
| chr10_+_128139191 | 0.02 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr10_+_58091287 | 0.01 |
ENSMUST00000057659.14
ENSMUST00000162041.8 ENSMUST00000162860.8 |
Gcc2
|
GRIP and coiled-coil domain containing 2 |
| chr4_+_118266526 | 0.01 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr12_+_112555656 | 0.01 |
ENSMUST00000220786.2
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr16_-_23339329 | 0.01 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr4_+_98435155 | 0.01 |
ENSMUST00000030290.8
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr6_+_42411469 | 0.01 |
ENSMUST00000059534.5
|
Tas2r126
|
taste receptor, type 2, member 126 |
| chr2_+_162829250 | 0.00 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr4_+_98434930 | 0.00 |
ENSMUST00000102792.10
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr10_-_14581203 | 0.00 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chrX_+_10118544 | 0.00 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr11_+_98337655 | 0.00 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr1_+_139429430 | 0.00 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chrX_+_10118600 | 0.00 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr4_+_101766318 | 0.00 |
ENSMUST00000075999.4
|
Pramel18
|
PRAME like 18 |
| chr9_+_108270020 | 0.00 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr4_+_101797604 | 0.00 |
ENSMUST00000052027.8
|
Pramel19
|
PRAME like 19 |
| chr9_+_58395850 | 0.00 |
ENSMUST00000085658.5
|
Insyn1
|
inhibitory synaptic factor 1 |
| chr16_+_22769844 | 0.00 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr2_+_3115250 | 0.00 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppard
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.7 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 1.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0071724 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.4 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.4 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.3 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |