Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
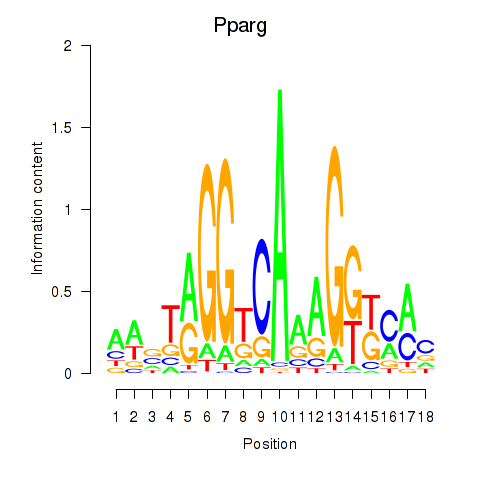
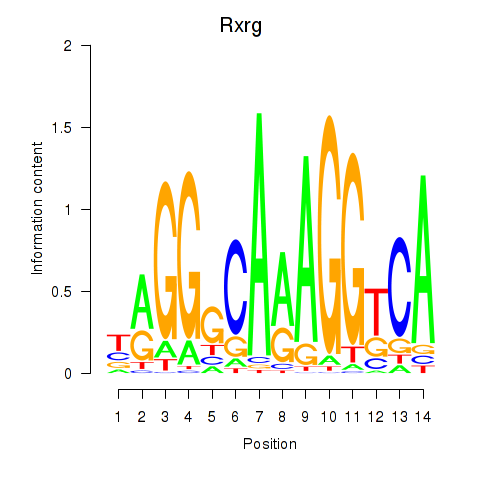
Results for Pparg_Rxrg
Z-value: 0.28
Transcription factors associated with Pparg_Rxrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pparg
|
ENSMUSG00000000440.13 | peroxisome proliferator activated receptor gamma |
|
Rxrg
|
ENSMUSG00000015843.11 | retinoid X receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pparg | mm39_v1_chr6_+_115398996_115399030 | 0.68 | 2.1e-01 | Click! |
| Rxrg | mm39_v1_chr1_+_167426019_167426049 | -0.66 | 2.3e-01 | Click! |
Activity profile of Pparg_Rxrg motif
Sorted Z-values of Pparg_Rxrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_13076024 | 0.29 |
ENSMUST00000033820.4
|
F7
|
coagulation factor VII |
| chr6_-_124790029 | 0.22 |
ENSMUST00000149610.3
|
Tpi1
|
triosephosphate isomerase 1 |
| chr4_+_150321142 | 0.21 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr11_-_69906171 | 0.18 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr17_+_26882171 | 0.17 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr19_+_4644365 | 0.14 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr11_+_52265090 | 0.14 |
ENSMUST00000020673.3
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr4_+_152270510 | 0.14 |
ENSMUST00000167926.8
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr5_-_115257336 | 0.14 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr9_-_15212849 | 0.14 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr7_+_43093507 | 0.13 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr4_+_152270636 | 0.13 |
ENSMUST00000030779.10
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr11_-_118292678 | 0.13 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr18_+_60880149 | 0.13 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr9_-_55419442 | 0.12 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr11_+_87684548 | 0.12 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr14_-_56339915 | 0.11 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr11_-_4045343 | 0.11 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr10_-_34003933 | 0.11 |
ENSMUST00000062784.8
|
Calhm6
|
calcium homeostasis modulator family member 6 |
| chr17_+_48606948 | 0.10 |
ENSMUST00000233092.2
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr6_-_90302159 | 0.10 |
ENSMUST00000070890.6
|
Chst13
|
carbohydrate sulfotransferase 13 |
| chr6_+_72575458 | 0.10 |
ENSMUST00000070597.13
ENSMUST00000176364.8 ENSMUST00000176168.3 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr3_-_88410495 | 0.10 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr9_+_43957241 | 0.10 |
ENSMUST00000214627.2
|
Thy1
|
thymus cell antigen 1, theta |
| chr19_-_4675352 | 0.10 |
ENSMUST00000224707.2
ENSMUST00000237059.2 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr2_-_32594156 | 0.10 |
ENSMUST00000127812.3
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr15_-_37008011 | 0.10 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr11_+_87684299 | 0.10 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr8_-_58106057 | 0.09 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr5_-_134205559 | 0.09 |
ENSMUST00000076228.3
|
Rcc1l
|
reculator of chromosome condensation 1 like |
| chr9_+_43956969 | 0.09 |
ENSMUST00000215809.2
|
Thy1
|
thymus cell antigen 1, theta |
| chr6_+_7844759 | 0.09 |
ENSMUST00000040159.6
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr5_-_110927803 | 0.09 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr15_+_101184488 | 0.09 |
ENSMUST00000229525.2
ENSMUST00000230525.2 |
Atg101
|
autophagy related 101 |
| chr8_-_58106027 | 0.09 |
ENSMUST00000110316.3
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr14_+_31216305 | 0.09 |
ENSMUST00000022446.7
|
Eaf1
|
ELL associated factor 1 |
| chr4_+_43506966 | 0.09 |
ENSMUST00000030183.10
|
Car9
|
carbonic anhydrase 9 |
| chr19_-_60862964 | 0.09 |
ENSMUST00000025961.7
|
Prdx3
|
peroxiredoxin 3 |
| chr7_+_120633639 | 0.09 |
ENSMUST00000033163.8
|
Mettl9
|
methyltransferase like 9 |
| chr3_+_94600863 | 0.09 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr10_-_35587888 | 0.08 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr17_-_35100980 | 0.08 |
ENSMUST00000152417.8
ENSMUST00000146299.8 |
C2
Gm20547
|
complement component 2 (within H-2S) predicted gene 20547 |
| chr3_-_58433313 | 0.08 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr5_-_105387395 | 0.08 |
ENSMUST00000065588.7
|
Gbp10
|
guanylate-binding protein 10 |
| chr7_+_46490899 | 0.08 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr19_-_6899121 | 0.08 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr10_+_128626772 | 0.08 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr16_+_33614715 | 0.08 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr4_+_152284261 | 0.08 |
ENSMUST00000105652.3
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr4_-_108690741 | 0.08 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr5_-_110928436 | 0.08 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr2_-_77110933 | 0.08 |
ENSMUST00000102659.2
|
Sestd1
|
SEC14 and spectrin domains 1 |
| chr10_-_119075910 | 0.08 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr11_-_88754543 | 0.07 |
ENSMUST00000107904.3
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr10_+_85665234 | 0.07 |
ENSMUST00000217667.2
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr7_-_29931612 | 0.07 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chrM_+_8603 | 0.07 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr2_-_164197987 | 0.07 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr4_+_97665843 | 0.07 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr17_+_35268942 | 0.07 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr12_+_104180795 | 0.07 |
ENSMUST00000121337.8
ENSMUST00000167049.8 ENSMUST00000101080.2 |
Serpina3f
|
serine (or cysteine) peptidase inhibitor, clade A, member 3F |
| chr7_-_117728790 | 0.07 |
ENSMUST00000206491.2
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr2_+_92205651 | 0.07 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr19_+_10019023 | 0.07 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr10_+_78410180 | 0.07 |
ENSMUST00000218061.2
ENSMUST00000218787.2 ENSMUST00000105384.5 ENSMUST00000218875.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr9_+_83807162 | 0.07 |
ENSMUST00000190637.7
ENSMUST00000034801.11 |
Bckdhb
|
branched chain ketoacid dehydrogenase E1, beta polypeptide |
| chr9_-_103105638 | 0.07 |
ENSMUST00000126359.2
|
Trf
|
transferrin |
| chr4_-_135699205 | 0.07 |
ENSMUST00000105852.8
|
Lypla2
|
lysophospholipase 2 |
| chr9_-_45923908 | 0.06 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr16_+_33614378 | 0.06 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr7_-_126399208 | 0.06 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr12_-_79239022 | 0.06 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr3_+_93462387 | 0.06 |
ENSMUST00000045756.14
|
S100a10
|
S100 calcium binding protein A10 (calpactin) |
| chr5_-_137529251 | 0.06 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr16_+_4501934 | 0.06 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr19_+_5012362 | 0.06 |
ENSMUST00000236917.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr5_-_130053120 | 0.06 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr7_-_141936355 | 0.06 |
ENSMUST00000133843.8
|
Gm49369
|
predicted gene, 49369 |
| chr1_-_16174387 | 0.06 |
ENSMUST00000149566.2
|
Rpl7
|
ribosomal protein L7 |
| chr7_-_126399778 | 0.06 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr19_-_6899173 | 0.06 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr5_-_137530214 | 0.06 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr11_-_118292758 | 0.06 |
ENSMUST00000043722.10
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr18_+_74912268 | 0.06 |
ENSMUST00000041053.11
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chrX_+_149377416 | 0.06 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr4_+_141028539 | 0.06 |
ENSMUST00000006614.3
|
Epha2
|
Eph receptor A2 |
| chr11_+_23616477 | 0.06 |
ENSMUST00000143117.2
|
Pus10
|
pseudouridylate synthase 10 |
| chr3_-_105708601 | 0.06 |
ENSMUST00000197094.5
ENSMUST00000198004.2 |
Rap1a
|
RAS-related protein 1a |
| chr7_-_117728815 | 0.06 |
ENSMUST00000032888.10
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr6_-_115739284 | 0.06 |
ENSMUST00000166254.7
ENSMUST00000170625.8 |
Tmem40
|
transmembrane protein 40 |
| chr5_-_129916283 | 0.06 |
ENSMUST00000094280.4
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr9_-_20984790 | 0.06 |
ENSMUST00000010348.7
|
Fdx2
|
ferredoxin 2 |
| chr5_+_115417725 | 0.06 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr14_+_122771734 | 0.06 |
ENSMUST00000154206.8
ENSMUST00000038374.13 ENSMUST00000135578.8 |
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide |
| chr1_-_120047868 | 0.05 |
ENSMUST00000112648.8
|
Dbi
|
diazepam binding inhibitor |
| chr4_+_95855442 | 0.05 |
ENSMUST00000030306.14
|
Hook1
|
hook microtubule tethering protein 1 |
| chr9_+_107852733 | 0.05 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr16_+_20492267 | 0.05 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr9_+_21914513 | 0.05 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr10_+_78410803 | 0.05 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr19_+_4022322 | 0.05 |
ENSMUST00000143380.3
|
Aldh3b2
|
aldehyde dehydrogenase 3 family, member B2 |
| chr19_-_6168518 | 0.05 |
ENSMUST00000113533.3
|
Sac3d1
|
SAC3 domain containing 1 |
| chr7_+_140659672 | 0.05 |
ENSMUST00000066873.5
ENSMUST00000163041.2 |
Pkp3
|
plakophilin 3 |
| chr7_-_102214636 | 0.05 |
ENSMUST00000106913.3
ENSMUST00000033264.12 |
Trim21
|
tripartite motif-containing 21 |
| chr14_-_56499690 | 0.05 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr17_+_35069347 | 0.05 |
ENSMUST00000097343.11
ENSMUST00000173357.8 ENSMUST00000173065.8 ENSMUST00000165953.3 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr7_-_126399574 | 0.05 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr12_+_55276953 | 0.05 |
ENSMUST00000218879.2
|
Srp54c
|
signal recognition particle 54C |
| chr5_+_67464284 | 0.05 |
ENSMUST00000113676.6
ENSMUST00000162372.8 |
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr14_+_8508484 | 0.05 |
ENSMUST00000065865.10
ENSMUST00000225891.2 |
Thoc7
|
THO complex 7 |
| chr4_+_118286898 | 0.05 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr7_-_19449319 | 0.05 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr19_+_11493825 | 0.05 |
ENSMUST00000163078.8
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr3_+_115681486 | 0.05 |
ENSMUST00000189799.7
ENSMUST00000200258.2 |
Dph5
|
diphthamide biosynthesis 5 |
| chr18_-_36859732 | 0.05 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr14_-_54855446 | 0.05 |
ENSMUST00000227257.2
ENSMUST00000022803.6 |
Psmb5
|
proteasome (prosome, macropain) subunit, beta type 5 |
| chr8_+_85415935 | 0.05 |
ENSMUST00000125370.10
ENSMUST00000175784.2 |
Trmt1
|
tRNA methyltransferase 1 |
| chr8_+_124170027 | 0.05 |
ENSMUST00000108830.2
|
Def8
|
differentially expressed in FDCP 8 |
| chr13_-_24945844 | 0.05 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr2_+_180102772 | 0.05 |
ENSMUST00000038225.8
|
Slco4a1
|
solute carrier organic anion transporter family, member 4a1 |
| chr13_-_55676334 | 0.05 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr3_-_100876960 | 0.05 |
ENSMUST00000076941.12
|
Ttf2
|
transcription termination factor, RNA polymerase II |
| chr2_-_164198427 | 0.05 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr11_+_96209093 | 0.05 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr3_-_107925122 | 0.05 |
ENSMUST00000126593.3
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr7_-_4869160 | 0.05 |
ENSMUST00000064547.13
|
Isoc2b
|
isochorismatase domain containing 2b |
| chr7_+_43086432 | 0.04 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr5_+_36622342 | 0.04 |
ENSMUST00000031099.4
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr4_+_118266582 | 0.04 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr15_-_78377926 | 0.04 |
ENSMUST00000163494.3
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr9_-_20871081 | 0.04 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr10_+_82669785 | 0.04 |
ENSMUST00000219368.3
|
Txnrd1
|
thioredoxin reductase 1 |
| chr6_-_124689094 | 0.04 |
ENSMUST00000004379.8
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr12_+_82216193 | 0.04 |
ENSMUST00000166429.9
ENSMUST00000220963.2 |
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr7_-_100661220 | 0.04 |
ENSMUST00000207916.2
|
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr11_+_4833186 | 0.04 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr10_+_78412783 | 0.04 |
ENSMUST00000219588.2
|
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr3_-_153650068 | 0.04 |
ENSMUST00000150070.2
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr10_+_43400074 | 0.04 |
ENSMUST00000057649.8
ENSMUST00000216543.2 |
Gm9803
|
predicted gene 9803 |
| chr7_+_28533279 | 0.04 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr2_+_154498917 | 0.04 |
ENSMUST00000044277.10
|
Chmp4b
|
charged multivesicular body protein 4B |
| chr6_-_119365632 | 0.04 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr10_+_61556371 | 0.04 |
ENSMUST00000080099.6
|
Aifm2
|
apoptosis-inducing factor, mitochondrion-associated 2 |
| chr17_+_64244946 | 0.04 |
ENSMUST00000038080.7
|
Fer
|
fer (fms/fps related) protein kinase |
| chr19_+_46695889 | 0.04 |
ENSMUST00000003655.9
|
As3mt
|
arsenite methyltransferase |
| chr5_+_31855009 | 0.04 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr2_+_34661982 | 0.04 |
ENSMUST00000028222.13
ENSMUST00000100171.3 |
Hspa5
|
heat shock protein 5 |
| chr12_-_13299197 | 0.04 |
ENSMUST00000071103.10
|
Ddx1
|
DEAD box helicase 1 |
| chr7_-_100661181 | 0.04 |
ENSMUST00000178340.3
ENSMUST00000037540.5 |
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr11_+_80320558 | 0.04 |
ENSMUST00000173565.2
|
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr3_+_89328926 | 0.04 |
ENSMUST00000094378.10
ENSMUST00000137793.2 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr11_-_98478018 | 0.04 |
ENSMUST00000052919.8
|
Ormdl3
|
ORM1-like 3 (S. cerevisiae) |
| chr11_-_69872050 | 0.04 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr6_+_85408953 | 0.04 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr18_-_35795233 | 0.04 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr5_-_45607485 | 0.04 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr5_-_45607463 | 0.04 |
ENSMUST00000197946.5
ENSMUST00000127562.3 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr2_-_32321116 | 0.04 |
ENSMUST00000127961.3
ENSMUST00000136361.8 ENSMUST00000052119.14 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr2_-_48839276 | 0.04 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr19_+_36532061 | 0.04 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr13_+_73476629 | 0.04 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr13_-_55677109 | 0.04 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr18_-_64649497 | 0.04 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr7_+_43086554 | 0.04 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr5_+_114924106 | 0.04 |
ENSMUST00000137519.2
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr4_-_155482516 | 0.04 |
ENSMUST00000030925.3
|
Gabrd
|
gamma-aminobutyric acid (GABA) A receptor, subunit delta |
| chr9_+_111011388 | 0.04 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_-_55127312 | 0.04 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr9_+_110173253 | 0.04 |
ENSMUST00000199709.3
|
Scap
|
SREBF chaperone |
| chr11_+_73090270 | 0.04 |
ENSMUST00000006105.7
|
Shpk
|
sedoheptulokinase |
| chr3_-_89905547 | 0.04 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr12_+_84332006 | 0.04 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr19_+_5012396 | 0.04 |
ENSMUST00000235229.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr9_+_59563872 | 0.04 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr2_+_3771709 | 0.04 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr14_+_121148625 | 0.04 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr4_+_128887017 | 0.04 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chr19_+_4644425 | 0.04 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr19_+_24853039 | 0.04 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr14_+_52348396 | 0.04 |
ENSMUST00000111600.11
|
Rpgrip1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr7_+_140658101 | 0.04 |
ENSMUST00000106039.9
|
Pkp3
|
plakophilin 3 |
| chr11_-_58829738 | 0.04 |
ENSMUST00000094151.6
|
Rnf187
|
ring finger protein 187 |
| chr6_+_129510117 | 0.04 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr12_-_108859123 | 0.04 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr3_-_88241789 | 0.04 |
ENSMUST00000001456.11
|
Tmem79
|
transmembrane protein 79 |
| chr8_+_71995554 | 0.04 |
ENSMUST00000034272.9
|
Mvb12a
|
multivesicular body subunit 12A |
| chr3_-_153650269 | 0.04 |
ENSMUST00000072697.13
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr14_+_59438658 | 0.04 |
ENSMUST00000173547.8
ENSMUST00000043227.13 ENSMUST00000022551.14 |
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr7_-_12787611 | 0.04 |
ENSMUST00000182490.2
|
Mzf1
|
myeloid zinc finger 1 |
| chr17_+_34823790 | 0.04 |
ENSMUST00000173242.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr19_-_4062738 | 0.04 |
ENSMUST00000136921.2
ENSMUST00000042497.14 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr7_+_37882642 | 0.04 |
ENSMUST00000178207.10
ENSMUST00000179525.10 |
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr2_+_127112127 | 0.04 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr4_+_41760454 | 0.04 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr15_-_76422928 | 0.04 |
ENSMUST00000023219.9
|
Fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr12_+_72583114 | 0.04 |
ENSMUST00000044352.8
|
Pcnx4
|
pecanex homolog 4 |
| chr1_-_75193937 | 0.04 |
ENSMUST00000186971.2
ENSMUST00000188593.2 |
Tuba4a
|
tubulin, alpha 4A |
| chr7_+_19248345 | 0.04 |
ENSMUST00000135972.2
|
Trappc6a
|
trafficking protein particle complex 6A |
| chr17_-_45903410 | 0.04 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr4_+_149569672 | 0.04 |
ENSMUST00000124413.8
ENSMUST00000141293.8 |
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pparg_Rxrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.2 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.2 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.0 | 0.3 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.0 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.0 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.1 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.0 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:1990796 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0071920 | cleavage body(GO:0071920) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.0 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.0 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |