Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
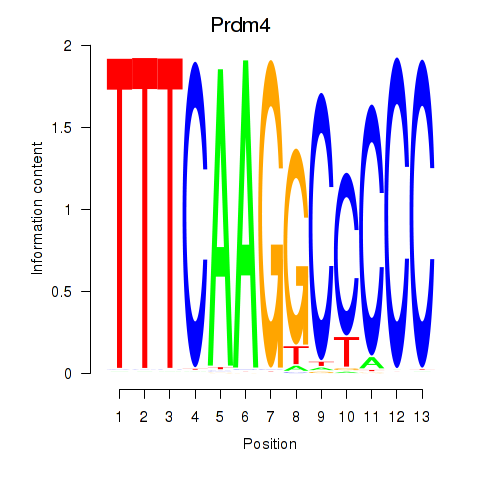
Results for Prdm4
Z-value: 0.67
Transcription factors associated with Prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm4
|
ENSMUSG00000035529.11 | PR domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm4 | mm39_v1_chr10_-_85752932_85753002 | 0.76 | 1.4e-01 | Click! |
Activity profile of Prdm4 motif
Sorted Z-values of Prdm4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23735822 | 1.17 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr5_-_142803135 | 0.82 |
ENSMUST00000198181.2
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr12_+_113115632 | 0.74 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr19_-_4213347 | 0.38 |
ENSMUST00000025749.15
|
Rps6kb2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr10_+_126814542 | 0.37 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr5_-_142803405 | 0.17 |
ENSMUST00000151477.8
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr2_+_97298002 | 0.09 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr7_-_30443106 | 0.09 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr15_-_99372575 | 0.04 |
ENSMUST00000040313.6
|
Bcdin3d
|
BCDIN3 domain containing |
| chr19_-_12080698 | 0.01 |
ENSMUST00000216069.2
ENSMUST00000215374.2 |
Olfr1427
|
olfactory receptor 1427 |
| chrX_+_135828069 | 0.01 |
ENSMUST00000059808.5
|
H2bw2
|
H2B.W histone 2 |
| chrX_+_21581135 | 0.01 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr11_-_70423765 | 0.01 |
ENSMUST00000209971.2
|
Gm40193
|
predicted gene, 40193 |
| chr11_-_43727071 | 0.00 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr15_-_98093423 | 0.00 |
ENSMUST00000165379.8
|
Olfr288
|
olfactory receptor 288 |
| chr8_+_123920682 | 0.00 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr7_+_43616163 | 0.00 |
ENSMUST00000078835.3
|
Klk1b1
|
kallikrein 1-related peptidase b1 |
| chr9_+_106503781 | 0.00 |
ENSMUST00000171091.4
|
Iqcf6
|
IQ motif containing F6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.7 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |