Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Prop1
Z-value: 0.57
Transcription factors associated with Prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prop1
|
ENSMUSG00000044542.4 | paired like homeodomain factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prop1 | mm39_v1_chr11_-_50844572_50844596 | 0.89 | 4.3e-02 | Click! |
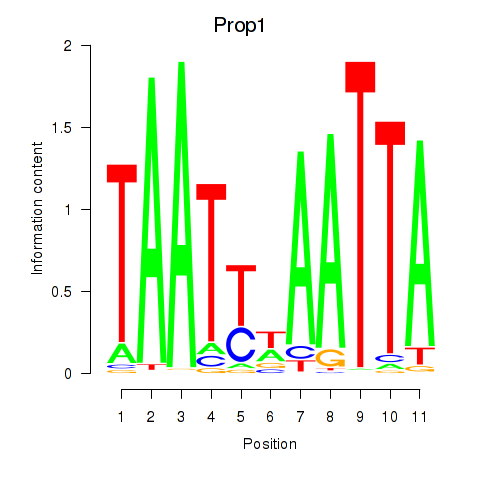
Activity profile of Prop1 motif
Sorted Z-values of Prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_57340712 | 0.72 |
ENSMUST00000228156.2
ENSMUST00000227186.2 ENSMUST00000228294.2 ENSMUST00000228342.2 |
Vmn1r17
|
vomeronasal 1 receptor 17 |
| chr2_+_88470886 | 0.51 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr9_+_38491474 | 0.49 |
ENSMUST00000217160.3
|
Olfr912
|
olfactory receptor 912 |
| chr16_+_65317389 | 0.44 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr2_+_111084861 | 0.40 |
ENSMUST00000218065.2
|
Olfr1276
|
olfactory receptor 1276 |
| chr11_-_79414542 | 0.38 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b |
| chr7_-_85065128 | 0.37 |
ENSMUST00000171213.3
ENSMUST00000233336.2 |
Vmn2r69
|
vomeronasal 2, receptor 69 |
| chr9_+_39932760 | 0.36 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr5_+_123280250 | 0.36 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr11_+_73851643 | 0.34 |
ENSMUST00000213134.2
ENSMUST00000216291.2 |
Olfr397
|
olfactory receptor 397 |
| chr2_+_87696836 | 0.32 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr11_+_101932328 | 0.32 |
ENSMUST00000123895.8
ENSMUST00000017453.12 ENSMUST00000107163.9 ENSMUST00000107164.3 |
Cd300lg
|
CD300 molecule like family member G |
| chr7_+_102954855 | 0.32 |
ENSMUST00000214577.2
|
Olfr596
|
olfactory receptor 596 |
| chr2_-_88768449 | 0.31 |
ENSMUST00000215205.2
ENSMUST00000213412.2 |
Olfr1211
|
olfactory receptor 1211 |
| chr6_+_57180275 | 0.30 |
ENSMUST00000226892.2
ENSMUST00000227421.2 |
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr18_+_32200781 | 0.30 |
ENSMUST00000025243.5
ENSMUST00000212675.2 |
Iws1
|
IWS1, SUPT6 interacting protein |
| chr2_+_87185159 | 0.29 |
ENSMUST00000215163.3
|
Olfr1120
|
olfactory receptor 1120 |
| chr7_-_85525474 | 0.29 |
ENSMUST00000077478.8
ENSMUST00000233729.2 ENSMUST00000233079.2 ENSMUST00000233315.2 ENSMUST00000233115.2 ENSMUST00000233748.2 ENSMUST00000232965.2 ENSMUST00000232897.2 |
Vmn2r73
|
vomeronasal 2, receptor 73 |
| chr6_+_57405915 | 0.29 |
ENSMUST00000227909.2
|
Vmn1r20
|
vomeronasal 1 receptor 20 |
| chr2_-_86926352 | 0.29 |
ENSMUST00000217066.3
ENSMUST00000214636.2 |
Olfr1109
|
olfactory receptor 1109 |
| chr13_+_65285903 | 0.29 |
ENSMUST00000206324.3
|
Olfr465-ps1
|
olfactory receptor 465, pseudogene 1 |
| chr18_+_37453427 | 0.28 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr9_-_49710058 | 0.28 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr6_-_97408367 | 0.28 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chr2_+_88285211 | 0.27 |
ENSMUST00000219871.2
ENSMUST00000213115.3 |
Olfr1183
|
olfactory receptor 1183 |
| chrX_+_109857866 | 0.27 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr12_+_119356318 | 0.27 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chrX_+_158491589 | 0.26 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_-_102998876 | 0.26 |
ENSMUST00000215042.2
|
Olfr600
|
olfactory receptor 600 |
| chr2_-_85966272 | 0.25 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr9_-_39618413 | 0.25 |
ENSMUST00000215192.2
|
Olfr149
|
olfactory receptor 149 |
| chr6_-_70237939 | 0.25 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr9_-_14651985 | 0.24 |
ENSMUST00000076946.4
ENSMUST00000115644.10 |
Piwil4
|
piwi-like RNA-mediated gene silencing 4 |
| chr5_-_62923463 | 0.24 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_+_84500854 | 0.24 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr4_+_100336003 | 0.24 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr16_-_4698148 | 0.24 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr2_-_86008164 | 0.23 |
ENSMUST00000215171.2
|
Olfr1044
|
olfactory receptor 1044 |
| chr17_+_28945057 | 0.23 |
ENSMUST00000233003.2
|
Gm4356
|
predicted gene 4356 |
| chr4_-_132191181 | 0.23 |
ENSMUST00000102567.4
|
Med18
|
mediator complex subunit 18 |
| chr9_-_49710190 | 0.22 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr7_-_85610837 | 0.22 |
ENSMUST00000232886.2
ENSMUST00000232758.2 ENSMUST00000166355.3 |
Vmn2r74
|
vomeronasal 2, receptor 74 |
| chr7_-_11414074 | 0.22 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr13_+_49658249 | 0.22 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr6_-_58195806 | 0.22 |
ENSMUST00000228530.2
ENSMUST00000226666.2 |
Vmn1r27
|
vomeronasal 1 receptor 27 |
| chrM_+_2743 | 0.21 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr7_+_107649900 | 0.20 |
ENSMUST00000214599.2
ENSMUST00000209805.3 |
Olfr479
|
olfactory receptor 479 |
| chr2_-_89491811 | 0.20 |
ENSMUST00000215730.3
|
Olfr1250
|
olfactory receptor 1250 |
| chr19_+_13594739 | 0.20 |
ENSMUST00000217061.3
ENSMUST00000209005.4 ENSMUST00000208347.3 |
Olfr1487
|
olfactory receptor 1487 |
| chr2_-_45000389 | 0.20 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_158623460 | 0.20 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_+_124114504 | 0.19 |
ENSMUST00000058994.6
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr7_-_103543692 | 0.19 |
ENSMUST00000081748.6
|
Olfr64
|
olfactory receptor 64 |
| chr17_+_37769807 | 0.19 |
ENSMUST00000214668.2
ENSMUST00000217602.2 ENSMUST00000214938.2 |
Olfr109
|
olfactory receptor 109 |
| chr9_+_56979307 | 0.19 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr18_-_9282754 | 0.18 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chr2_-_88581690 | 0.18 |
ENSMUST00000215179.3
ENSMUST00000215529.3 |
Olfr1198
|
olfactory receptor 1198 |
| chr2_-_88848668 | 0.17 |
ENSMUST00000216000.2
ENSMUST00000217000.2 |
Olfr1216
|
olfactory receptor 1216 |
| chr2_-_36782948 | 0.17 |
ENSMUST00000217215.3
|
Olfr353
|
olfactory receptor 353 |
| chr3_+_59832635 | 0.16 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr3_-_146200870 | 0.15 |
ENSMUST00000093951.3
|
Spata1
|
spermatogenesis associated 1 |
| chr7_+_84502761 | 0.15 |
ENSMUST00000217039.3
ENSMUST00000211582.2 |
Olfr291
|
olfactory receptor 291 |
| chr2_-_89671899 | 0.15 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr17_+_18801989 | 0.15 |
ENSMUST00000165692.8
|
Vmn2r96
|
vomeronasal 2, receptor 96 |
| chr7_-_126574959 | 0.15 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr7_-_86016045 | 0.15 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr7_+_6470100 | 0.14 |
ENSMUST00000207658.4
ENSMUST00000215302.3 |
Olfr1346
|
olfactory receptor 1346 |
| chr6_-_57991216 | 0.14 |
ENSMUST00000228951.2
|
Vmn1r26
|
vomeronasal 1 receptor 26 |
| chr2_+_61423469 | 0.14 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr13_+_83723743 | 0.14 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr12_-_56581823 | 0.13 |
ENSMUST00000178477.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr16_+_62635039 | 0.13 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chrX_-_99638466 | 0.13 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr4_-_3938352 | 0.13 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chrM_+_11735 | 0.13 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr9_+_100479732 | 0.13 |
ENSMUST00000124487.8
|
Stag1
|
stromal antigen 1 |
| chr8_+_72837021 | 0.13 |
ENSMUST00000213940.2
|
Olfr373
|
olfactory receptor 373 |
| chr14_-_50479161 | 0.12 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr1_+_130754413 | 0.12 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr11_-_113574981 | 0.12 |
ENSMUST00000120194.2
|
Fam104a
|
family with sequence similarity 104, member A |
| chr14_+_84680993 | 0.12 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr2_-_45001141 | 0.12 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_+_3993774 | 0.11 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr11_-_46057224 | 0.11 |
ENSMUST00000020679.3
|
Nipal4
|
NIPA-like domain containing 4 |
| chr2_+_83554770 | 0.11 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr9_+_77959206 | 0.11 |
ENSMUST00000024104.9
|
Gcm1
|
glial cells missing homolog 1 |
| chr2_+_61423421 | 0.11 |
ENSMUST00000112495.8
ENSMUST00000112501.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr7_+_104730412 | 0.10 |
ENSMUST00000215704.2
ENSMUST00000214328.3 |
Olfr679
|
olfactory receptor 679 |
| chrX_+_56008685 | 0.10 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr2_+_88479038 | 0.10 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1 |
| chr13_+_65298054 | 0.10 |
ENSMUST00000214214.2
|
Olfr466
|
olfactory receptor 466 |
| chr2_-_88157559 | 0.10 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr4_+_35152056 | 0.10 |
ENSMUST00000058595.7
|
Ifnk
|
interferon kappa |
| chr7_-_104677667 | 0.10 |
ENSMUST00000215899.2
ENSMUST00000214318.3 |
Olfr675
|
olfactory receptor 675 |
| chr7_+_79939747 | 0.10 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr8_-_3675525 | 0.10 |
ENSMUST00000144977.2
ENSMUST00000136105.8 ENSMUST00000128566.8 |
Pcp2
|
Purkinje cell protein 2 (L7) |
| chrX_-_58211440 | 0.10 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr7_-_12829100 | 0.09 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr2_-_36899347 | 0.09 |
ENSMUST00000216437.2
|
Olfr358
|
olfactory receptor 358 |
| chr8_-_65582206 | 0.09 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr9_-_39920878 | 0.09 |
ENSMUST00000216463.3
|
Olfr980
|
olfactory receptor 980 |
| chr2_-_88913831 | 0.09 |
ENSMUST00000217421.2
ENSMUST00000214442.2 ENSMUST00000215225.3 |
Olfr1219
|
olfactory receptor 1219 |
| chr12_+_9080014 | 0.09 |
ENSMUST00000219488.2
ENSMUST00000219470.2 |
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr2_+_87854404 | 0.09 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr6_-_119925387 | 0.09 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr10_-_45346297 | 0.09 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr2_+_25596751 | 0.08 |
ENSMUST00000061483.9
|
Bmyc
|
brain expressed myelocytomatosis oncogene |
| chr5_+_31771281 | 0.08 |
ENSMUST00000031024.14
ENSMUST00000202033.2 |
Mrpl33
Gm43809
|
mitochondrial ribosomal protein L33 predicted gene 43809 |
| chr4_+_47386216 | 0.08 |
ENSMUST00000107725.3
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr10_+_129306867 | 0.08 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788 |
| chr18_+_69726055 | 0.08 |
ENSMUST00000114978.9
|
Tcf4
|
transcription factor 4 |
| chr2_-_86317730 | 0.08 |
ENSMUST00000217292.3
|
Olfr228
|
olfactory receptor 228 |
| chr18_+_69726431 | 0.08 |
ENSMUST00000201091.4
ENSMUST00000201037.4 ENSMUST00000114977.5 |
Tcf4
|
transcription factor 4 |
| chr16_-_64591509 | 0.08 |
ENSMUST00000076991.7
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr10_+_79832313 | 0.08 |
ENSMUST00000132517.8
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr19_-_13828056 | 0.08 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr16_+_56024676 | 0.08 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr13_+_95012107 | 0.07 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr14_+_73411249 | 0.07 |
ENSMUST00000166875.2
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr6_-_70412460 | 0.07 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr5_+_42225303 | 0.07 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr6_-_136150491 | 0.07 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr13_+_49761506 | 0.07 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr9_+_119939414 | 0.07 |
ENSMUST00000035106.12
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr4_+_115696388 | 0.07 |
ENSMUST00000019677.12
ENSMUST00000144427.8 ENSMUST00000106513.10 ENSMUST00000130819.8 ENSMUST00000151203.8 ENSMUST00000140315.2 |
Mknk1
|
MAP kinase-interacting serine/threonine kinase 1 |
| chr15_+_98350469 | 0.07 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr7_-_107856399 | 0.07 |
ENSMUST00000211508.3
|
Olfr488
|
olfactory receptor 488 |
| chr17_+_56920389 | 0.07 |
ENSMUST00000080492.7
|
Rpl36
|
ribosomal protein L36 |
| chr12_+_79344119 | 0.07 |
ENSMUST00000079533.12
|
Rad51b
|
RAD51 paralog B |
| chr10_-_63926044 | 0.07 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr6_-_3399451 | 0.07 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr6_-_82916655 | 0.07 |
ENSMUST00000000641.15
|
Sema4f
|
sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain |
| chr14_-_31139617 | 0.07 |
ENSMUST00000100730.10
|
Sh3bp5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr2_+_152528955 | 0.06 |
ENSMUST00000062148.9
|
Mcts2
|
malignant T cell amplified sequence 2 |
| chr17_-_25394445 | 0.06 |
ENSMUST00000224277.2
|
Percc1
|
proline and glutamate rich with coiled coil 1 |
| chr11_-_87783073 | 0.06 |
ENSMUST00000213672.2
ENSMUST00000213928.2 |
Olfr462
|
olfactory receptor 462 |
| chr7_-_6733411 | 0.06 |
ENSMUST00000239104.2
ENSMUST00000051209.11 |
Peg3
|
paternally expressed 3 |
| chr4_+_125940678 | 0.06 |
ENSMUST00000030675.8
|
Mrps15
|
mitochondrial ribosomal protein S15 |
| chr7_+_19725390 | 0.06 |
ENSMUST00000207805.2
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr7_-_108484213 | 0.06 |
ENSMUST00000217803.2
|
Olfr518
|
olfactory receptor 518 |
| chr3_-_116506345 | 0.06 |
ENSMUST00000169530.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr19_-_12093187 | 0.05 |
ENSMUST00000208391.3
ENSMUST00000214103.2 |
Olfr1428
|
olfactory receptor 1428 |
| chr3_+_41697046 | 0.05 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr1_-_85526517 | 0.05 |
ENSMUST00000093508.7
|
Sp110
|
Sp110 nuclear body protein |
| chr9_+_118307250 | 0.05 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr4_+_41348996 | 0.05 |
ENSMUST00000108060.10
ENSMUST00000072866.12 |
Ubap1
|
ubiquitin-associated protein 1 |
| chr9_-_48876290 | 0.05 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr10_+_129907079 | 0.05 |
ENSMUST00000216879.2
|
Olfr822
|
olfactory receptor 822 |
| chr2_+_158508609 | 0.05 |
ENSMUST00000103116.10
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr16_-_16962279 | 0.05 |
ENSMUST00000232033.2
ENSMUST00000231597.2 ENSMUST00000232540.2 |
Ccdc116
|
coiled-coil domain containing 116 |
| chr11_+_75542902 | 0.05 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr18_+_75133519 | 0.05 |
ENSMUST00000079716.6
|
Rpl17
|
ribosomal protein L17 |
| chr3_-_116506294 | 0.05 |
ENSMUST00000029569.9
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr5_+_9163244 | 0.05 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr17_-_54297657 | 0.05 |
ENSMUST00000024738.8
|
Sult1c1
|
sulfotransferase family, cytosolic, 1C, member 1 |
| chr17_+_37978659 | 0.05 |
ENSMUST00000216551.2
|
Olfr118
|
olfactory receptor 118 |
| chr11_+_62737936 | 0.05 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr18_+_38430015 | 0.05 |
ENSMUST00000236319.2
|
Rnf14
|
ring finger protein 14 |
| chr19_+_11514132 | 0.04 |
ENSMUST00000025581.7
|
Ms4a4d
|
membrane-spanning 4-domains, subfamily A, member 4D |
| chr18_-_55123153 | 0.04 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608 |
| chr4_-_88595161 | 0.04 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr7_+_106740521 | 0.04 |
ENSMUST00000210474.2
|
Olfr716
|
olfactory receptor 716 |
| chr2_-_85675827 | 0.04 |
ENSMUST00000213515.2
|
Olfr1019
|
olfactory receptor 1019 |
| chr13_-_3968157 | 0.04 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr16_-_94023976 | 0.04 |
ENSMUST00000227698.2
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| chr12_+_59176506 | 0.04 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr17_-_47922374 | 0.04 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr2_+_86655007 | 0.04 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr2_+_121787131 | 0.04 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr3_-_103698853 | 0.04 |
ENSMUST00000118317.8
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr3_-_41696906 | 0.04 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chrX_-_146212184 | 0.04 |
ENSMUST00000033646.9
|
Il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr11_-_46597885 | 0.04 |
ENSMUST00000055102.13
ENSMUST00000125008.2 |
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr1_+_167425953 | 0.03 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr11_-_54140462 | 0.03 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr11_-_59054107 | 0.03 |
ENSMUST00000069631.3
|
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr7_+_126895423 | 0.03 |
ENSMUST00000117762.8
|
Itgal
|
integrin alpha L |
| chr11_+_58643560 | 0.03 |
ENSMUST00000215071.2
|
Olfr316
|
olfactory receptor 316 |
| chr9_-_114811807 | 0.03 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr12_+_59176543 | 0.03 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr5_-_123804745 | 0.03 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr10_-_129630735 | 0.03 |
ENSMUST00000217283.2
ENSMUST00000214206.2 ENSMUST00000214878.2 |
Olfr810
|
olfactory receptor 810 |
| chr5_-_86475595 | 0.03 |
ENSMUST00000122377.2
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr2_-_87985537 | 0.03 |
ENSMUST00000216951.2
|
Olfr1167
|
olfactory receptor 1167 |
| chr19_-_23251179 | 0.03 |
ENSMUST00000087556.7
ENSMUST00000223934.2 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr4_+_146739384 | 0.03 |
ENSMUST00000070932.4
|
Zfp993
|
zinc finger protein 993 |
| chr6_-_70435020 | 0.03 |
ENSMUST00000198184.2
|
Igkv6-13
|
immunoglobulin kappa variable 6-13 |
| chr10_+_23727325 | 0.03 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr6_+_116466052 | 0.03 |
ENSMUST00000203700.3
|
Olfr211
|
olfactory receptor 211 |
| chr1_-_144651157 | 0.03 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr15_-_56557920 | 0.03 |
ENSMUST00000050544.8
|
Has2
|
hyaluronan synthase 2 |
| chr6_+_86986213 | 0.03 |
ENSMUST00000120240.8
|
Nfu1
|
NFU1 iron-sulfur cluster scaffold |
| chr3_-_79053182 | 0.03 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_-_25112256 | 0.03 |
ENSMUST00000200880.4
ENSMUST00000074040.4 |
Cxcl17
|
chemokine (C-X-C motif) ligand 17 |
| chr1_-_119432455 | 0.03 |
ENSMUST00000004565.15
|
Ralb
|
v-ral simian leukemia viral oncogene B |
| chr18_+_38429858 | 0.03 |
ENSMUST00000171461.3
ENSMUST00000235811.2 |
Rnf14
|
ring finger protein 14 |
| chr7_-_45343785 | 0.03 |
ENSMUST00000040636.9
|
Sec1
|
secretory blood group 1 |
| chr19_-_13807805 | 0.03 |
ENSMUST00000214475.2
ENSMUST00000067670.5 ENSMUST00000219674.2 ENSMUST00000216287.2 ENSMUST00000215760.2 |
Olfr1500
|
olfactory receptor 1500 |
| chr13_+_83723255 | 0.02 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_43046476 | 0.02 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.0 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.0 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |