Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
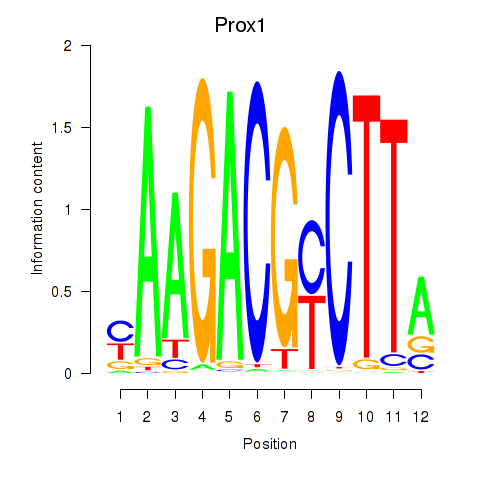
Results for Prox1
Z-value: 0.89
Transcription factors associated with Prox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prox1
|
ENSMUSG00000010175.14 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prox1 | mm39_v1_chr1_-_189902868_189902942 | -0.82 | 9.0e-02 | Click! |
Activity profile of Prox1 motif
Sorted Z-values of Prox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_67378886 | 0.73 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr2_+_36120438 | 0.61 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr13_+_9326513 | 0.56 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr4_-_133694607 | 0.50 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr19_-_15901919 | 0.50 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr19_-_8691797 | 0.48 |
ENSMUST00000206797.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr4_+_140428777 | 0.42 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr17_-_35827676 | 0.38 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr5_+_24305577 | 0.37 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr8_-_85807281 | 0.35 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr18_-_67378512 | 0.35 |
ENSMUST00000237631.2
|
Mppe1
|
metallophosphoesterase 1 |
| chr9_-_106124917 | 0.34 |
ENSMUST00000112524.9
ENSMUST00000219129.2 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr9_-_64633865 | 0.33 |
ENSMUST00000168366.2
|
Rab11a
|
RAB11A, member RAS oncogene family |
| chr5_-_124492734 | 0.31 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr8_-_85807308 | 0.30 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr19_+_41900360 | 0.29 |
ENSMUST00000011896.8
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr17_+_28059129 | 0.28 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr17_+_24645615 | 0.27 |
ENSMUST00000234304.2
ENSMUST00000024946.7 ENSMUST00000234150.2 |
Eci1
|
enoyl-Coenzyme A delta isomerase 1 |
| chr5_+_139238069 | 0.26 |
ENSMUST00000026976.12
|
Get4
|
golgi to ER traffic protein 4 |
| chr4_-_133694543 | 0.26 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr2_-_26800581 | 0.26 |
ENSMUST00000015920.12
ENSMUST00000139815.2 ENSMUST00000102899.10 |
Med22
|
mediator complex subunit 22 |
| chr5_+_124250360 | 0.24 |
ENSMUST00000024470.13
ENSMUST00000119269.6 ENSMUST00000196627.5 ENSMUST00000199457.5 ENSMUST00000198505.2 |
Ogfod2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr6_+_58617521 | 0.23 |
ENSMUST00000145161.8
ENSMUST00000203146.3 ENSMUST00000114294.8 ENSMUST00000204948.2 |
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr7_+_141995545 | 0.23 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr1_+_134487928 | 0.22 |
ENSMUST00000112198.3
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr7_+_28416194 | 0.21 |
ENSMUST00000032818.13
|
Fbxo17
|
F-box protein 17 |
| chr6_-_131365380 | 0.19 |
ENSMUST00000032309.13
ENSMUST00000087865.4 |
Ybx3
|
Y box protein 3 |
| chr5_+_139238089 | 0.19 |
ENSMUST00000130326.8
|
Get4
|
golgi to ER traffic protein 4 |
| chr17_+_29538157 | 0.19 |
ENSMUST00000114699.9
ENSMUST00000155348.3 ENSMUST00000234618.2 |
Pi16
|
peptidase inhibitor 16 |
| chr19_-_8691220 | 0.18 |
ENSMUST00000010239.6
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr12_+_111409087 | 0.18 |
ENSMUST00000109792.8
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr4_-_63779562 | 0.16 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr19_-_43512929 | 0.16 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr3_-_148696155 | 0.16 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr17_+_28059099 | 0.16 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr7_+_28416270 | 0.16 |
ENSMUST00000108279.9
|
Fbxo17
|
F-box protein 17 |
| chr18_-_12952925 | 0.15 |
ENSMUST00000119043.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr2_+_34296894 | 0.15 |
ENSMUST00000113123.8
|
Mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr14_-_59835285 | 0.14 |
ENSMUST00000022555.11
ENSMUST00000225839.2 ENSMUST00000056997.15 ENSMUST00000171683.3 ENSMUST00000167100.9 |
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr7_+_23781311 | 0.12 |
ENSMUST00000207002.2
ENSMUST00000068975.6 ENSMUST00000203854.3 |
Zfp180
|
zinc finger protein 180 |
| chr2_+_34296783 | 0.10 |
ENSMUST00000149383.8
ENSMUST00000124443.8 ENSMUST00000141253.2 ENSMUST00000113124.8 |
Mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr8_-_70929555 | 0.10 |
ENSMUST00000066597.13
ENSMUST00000210250.2 ENSMUST00000209415.2 ENSMUST00000166976.3 |
Klhl26
|
kelch-like 26 |
| chr17_-_35191127 | 0.09 |
ENSMUST00000087328.4
|
Hspa1a
|
heat shock protein 1A |
| chr11_+_115671523 | 0.09 |
ENSMUST00000239299.2
|
Tmem94
|
transmembrane protein 94 |
| chr8_+_31601837 | 0.09 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr2_-_136733282 | 0.09 |
ENSMUST00000028730.13
ENSMUST00000110089.9 ENSMUST00000227806.2 |
Mkks
ENSMUSG00000115423.3
|
McKusick-Kaufman syndrome novel protein |
| chr17_-_83821716 | 0.08 |
ENSMUST00000025095.9
ENSMUST00000167741.9 |
Cox7a2l
|
cytochrome c oxidase subunit 7A2 like |
| chr2_+_26800757 | 0.08 |
ENSMUST00000102898.5
|
Rpl7a
|
ribosomal protein L7A |
| chr11_+_19874403 | 0.08 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr3_+_20111958 | 0.07 |
ENSMUST00000002502.12
|
Hltf
|
helicase-like transcription factor |
| chr17_+_28059036 | 0.06 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr7_-_80597246 | 0.06 |
ENSMUST00000120285.8
|
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chrY_-_90850446 | 0.06 |
ENSMUST00000179623.2
|
Gm21748
|
predicted gene, 21748 |
| chr8_+_89015705 | 0.06 |
ENSMUST00000171456.9
|
Adcy7
|
adenylate cyclase 7 |
| chr11_-_115503704 | 0.06 |
ENSMUST00000106506.8
|
Mif4gd
|
MIF4G domain containing |
| chr7_-_126460820 | 0.05 |
ENSMUST00000129812.2
ENSMUST00000106342.8 |
Ino80e
|
INO80 complex subunit E |
| chr4_+_99717671 | 0.05 |
ENSMUST00000239518.1
ENSMUST00000239520.1 ENSMUST00000136874.4 |
Efcab7
|
EF-hand calcium binding domain 7 |
| chr8_+_61085890 | 0.05 |
ENSMUST00000160719.8
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr11_-_115824290 | 0.05 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr10_+_67371295 | 0.05 |
ENSMUST00000145754.8
ENSMUST00000145936.2 |
Egr2
|
early growth response 2 |
| chr1_+_78794475 | 0.04 |
ENSMUST00000057262.8
ENSMUST00000187432.2 |
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr11_+_4936824 | 0.04 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr1_+_134487893 | 0.04 |
ENSMUST00000047714.14
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr17_+_37356872 | 0.03 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr11_+_19874354 | 0.03 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr10_-_80649315 | 0.03 |
ENSMUST00000181039.8
ENSMUST00000180438.2 |
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr5_-_86780277 | 0.03 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr2_-_89159994 | 0.02 |
ENSMUST00000213860.2
ENSMUST00000215679.2 |
Olfr1232
|
olfactory receptor 1232 |
| chr1_+_161222980 | 0.02 |
ENSMUST00000028024.5
|
Tnfsf4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr14_+_43156329 | 0.02 |
ENSMUST00000228117.2
|
Gm9732
|
predicted gene 9732 |
| chr9_-_38614969 | 0.02 |
ENSMUST00000215612.2
|
Olfr919
|
olfactory receptor 919 |
| chr5_+_62968993 | 0.02 |
ENSMUST00000238474.2
|
Dthd1
|
death domain containing 1 |
| chr4_-_68872585 | 0.02 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr17_-_37574640 | 0.02 |
ENSMUST00000080759.5
|
Olfr98
|
olfactory receptor 98 |
| chr10_-_128969635 | 0.02 |
ENSMUST00000062314.4
|
Olfr770
|
olfactory receptor 770 |
| chr18_+_24786748 | 0.02 |
ENSMUST00000068006.9
|
Mocos
|
molybdenum cofactor sulfurase |
| chr18_-_68562385 | 0.01 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr14_-_56181993 | 0.01 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr4_+_130640611 | 0.01 |
ENSMUST00000156225.8
ENSMUST00000156742.8 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr14_+_53754386 | 0.01 |
ENSMUST00000103584.4
|
Trav6-6
|
T cell receptor alpha variable 6-6 |
| chr16_+_22965330 | 0.01 |
ENSMUST00000171309.2
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr15_-_86070338 | 0.01 |
ENSMUST00000044332.16
|
Cerk
|
ceramide kinase |
| chrX_-_42363663 | 0.01 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr14_+_50759296 | 0.01 |
ENSMUST00000215793.2
|
Olfr743
|
olfactory receptor 743 |
| chr2_-_168576155 | 0.01 |
ENSMUST00000109175.9
|
Atp9a
|
ATPase, class II, type 9A |
| chr3_+_153549846 | 0.01 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr9_-_7388047 | 0.01 |
ENSMUST00000047888.8
|
Mmp1b
|
matrix metallopeptidase 1b (interstitial collagenase) |
| chr11_-_59484115 | 0.01 |
ENSMUST00000215626.2
|
Olfr223
|
olfactory receptor 223 |
| chr7_+_127376267 | 0.01 |
ENSMUST00000144406.8
|
Setd1a
|
SET domain containing 1A |
| chr4_+_74160705 | 0.01 |
ENSMUST00000077851.10
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr7_+_107700056 | 0.01 |
ENSMUST00000215159.3
|
Olfr483
|
olfactory receptor 483 |
| chr14_+_42477895 | 0.01 |
ENSMUST00000100696.8
ENSMUST00000166527.2 |
Gm10378
|
predicted gene 10378 |
| chr14_+_42132167 | 0.01 |
ENSMUST00000089789.11
ENSMUST00000171967.2 |
Gm7995
|
predicted gene 7995 |
| chr3_+_92980844 | 0.01 |
ENSMUST00000195847.2
ENSMUST00000179064.2 |
Tdpoz8
|
TD and POZ domain containing 8 |
| chr4_-_62126126 | 0.01 |
ENSMUST00000220873.2
|
Zfp37
|
zinc finger protein 37 |
| chr4_-_137137088 | 0.01 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr7_-_48106037 | 0.01 |
ENSMUST00000188095.2
|
Mrgprb1
|
MAS-related GPR, member B1 |
| chr10_-_41487315 | 0.01 |
ENSMUST00000219054.2
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr6_-_73446560 | 0.01 |
ENSMUST00000070163.6
|
4931417E11Rik
|
RIKEN cDNA 4931417E11 gene |
| chr4_-_36951223 | 0.01 |
ENSMUST00000108122.8
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_-_169575203 | 0.01 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr12_-_83534482 | 0.00 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr18_-_32082624 | 0.00 |
ENSMUST00000064016.6
|
Gpr17
|
G protein-coupled receptor 17 |
| chr18_+_46983105 | 0.00 |
ENSMUST00000025358.4
ENSMUST00000234519.2 |
Lvrn
|
laeverin |
| chr3_-_121076745 | 0.00 |
ENSMUST00000135818.8
ENSMUST00000137234.2 |
Tlcd4
|
TLC domain containing 4 |
| chr18_+_89215438 | 0.00 |
ENSMUST00000237110.2
|
Cd226
|
CD226 antigen |
| chr5_+_95891653 | 0.00 |
ENSMUST00000136313.2
ENSMUST00000101011.4 |
Gm7982
|
predicted gene 7982 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.6 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.2 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.1 | 0.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 0.3 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0098837 | kinetochore microtubule(GO:0005828) postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.3 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |