Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
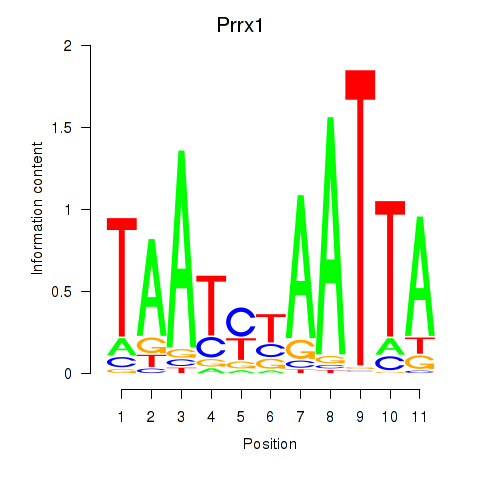
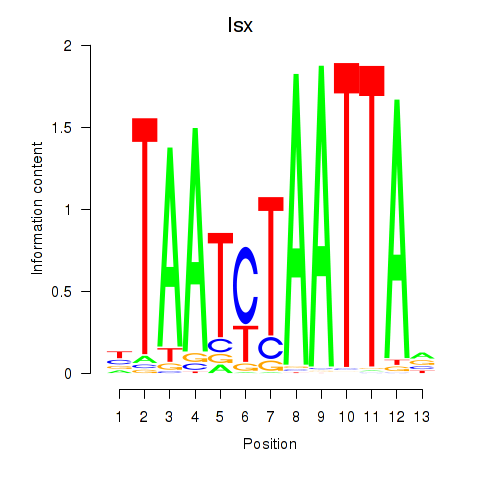
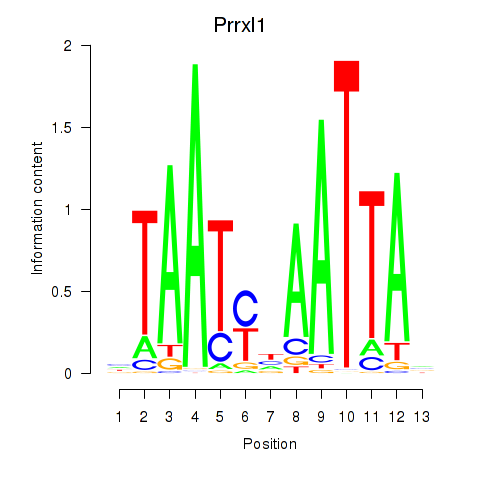
Results for Prrx1_Isx_Prrxl1
Z-value: 1.63
Transcription factors associated with Prrx1_Isx_Prrxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prrx1
|
ENSMUSG00000026586.17 | paired related homeobox 1 |
|
Isx
|
ENSMUSG00000031621.10 | intestine specific homeobox |
|
Prrxl1
|
ENSMUSG00000041730.15 | paired related homeobox protein-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prrxl1 | mm39_v1_chr14_+_32321341_32321498 | -0.87 | 5.5e-02 | Click! |
| Prrx1 | mm39_v1_chr1_-_163141230_163141246 | -0.79 | 1.1e-01 | Click! |
| Isx | mm39_v1_chr8_+_75599801_75599855 | -0.27 | 6.6e-01 | Click! |
Activity profile of Prrx1_Isx_Prrxl1 motif
Sorted Z-values of Prrx1_Isx_Prrxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_60907698 | 2.01 |
ENSMUST00000118551.8
|
Rps14
|
ribosomal protein S14 |
| chr8_+_106786190 | 1.82 |
ENSMUST00000109308.3
|
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr6_+_40619913 | 1.62 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr18_+_60907668 | 1.61 |
ENSMUST00000025511.11
|
Rps14
|
ribosomal protein S14 |
| chr4_-_87724533 | 1.57 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_-_34294377 | 1.41 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr18_-_43610829 | 1.36 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr16_+_49620883 | 1.35 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr1_-_24044688 | 1.24 |
ENSMUST00000027338.4
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr4_-_87724512 | 1.19 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr15_+_65682066 | 1.17 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chrX_+_37689503 | 0.97 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr3_+_159545309 | 0.93 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr3_+_32490300 | 0.91 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr10_+_82695013 | 0.90 |
ENSMUST00000020484.9
ENSMUST00000219962.3 |
Txnrd1
|
thioredoxin reductase 1 |
| chr15_+_100202061 | 0.78 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr12_-_79237722 | 0.78 |
ENSMUST00000085254.7
|
Rdh11
|
retinol dehydrogenase 11 |
| chr13_-_4659120 | 0.76 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr15_+_100202079 | 0.74 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr18_+_3382968 | 0.74 |
ENSMUST00000025073.12
|
Cul2
|
cullin 2 |
| chr11_-_117764258 | 0.70 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr8_-_58106057 | 0.70 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr15_+_100202021 | 0.69 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr12_-_75678092 | 0.69 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr7_+_99808452 | 0.68 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr19_-_24178000 | 0.68 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr5_+_52940391 | 0.65 |
ENSMUST00000031077.13
ENSMUST00000113904.9 ENSMUST00000199840.2 |
Zcchc4
|
zinc finger, CCHC domain containing 4 |
| chr17_+_64203017 | 0.64 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr10_+_99099084 | 0.62 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr14_-_104760051 | 0.62 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr12_-_73093953 | 0.61 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr8_-_58106027 | 0.59 |
ENSMUST00000110316.3
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr9_+_119170486 | 0.58 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr6_+_86342622 | 0.57 |
ENSMUST00000071492.9
|
Fam136a
|
family with sequence similarity 136, member A |
| chr17_-_36353582 | 0.57 |
ENSMUST00000058801.15
ENSMUST00000080015.12 ENSMUST00000077960.7 |
H2-T22
|
histocompatibility 2, T region locus 22 |
| chr11_-_17903861 | 0.53 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr13_+_96524802 | 0.53 |
ENSMUST00000099295.6
|
Poc5
|
POC5 centriolar protein |
| chr14_-_14255736 | 0.50 |
ENSMUST00000170111.3
|
Kctd6
|
potassium channel tetramerisation domain containing 6 |
| chr8_+_22682816 | 0.49 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr3_+_96552895 | 0.48 |
ENSMUST00000119365.8
ENSMUST00000029744.6 |
Itga10
|
integrin, alpha 10 |
| chr7_+_102090892 | 0.48 |
ENSMUST00000033283.10
|
Rrm1
|
ribonucleotide reductase M1 |
| chr12_-_84664001 | 0.47 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr9_-_96774719 | 0.47 |
ENSMUST00000154146.8
|
Pxylp1
|
2-phosphoxylose phosphatase 1 |
| chrM_+_7758 | 0.45 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr11_-_74615496 | 0.45 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr9_+_19533591 | 0.44 |
ENSMUST00000215372.2
|
Zfp317
|
zinc finger protein 317 |
| chr7_+_99808526 | 0.44 |
ENSMUST00000207825.2
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr14_-_36857202 | 0.44 |
ENSMUST00000165649.4
ENSMUST00000224769.2 |
Ghitm
|
growth hormone inducible transmembrane protein |
| chr1_-_160898181 | 0.44 |
ENSMUST00000035430.4
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr3_+_32490525 | 0.44 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chrM_+_7779 | 0.43 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr14_+_32507920 | 0.43 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr10_+_82695055 | 0.42 |
ENSMUST00000218694.2
|
Txnrd1
|
thioredoxin reductase 1 |
| chr7_+_126575781 | 0.42 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr16_+_38167352 | 0.40 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr12_-_80690573 | 0.40 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr9_+_118307412 | 0.40 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr6_-_86710250 | 0.39 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr7_-_24423715 | 0.39 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr17_+_46807637 | 0.39 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr11_+_58062467 | 0.39 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr9_+_119170360 | 0.38 |
ENSMUST00000039784.12
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr2_+_30331839 | 0.37 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr14_-_76248274 | 0.37 |
ENSMUST00000088922.5
|
Gtf2f2
|
general transcription factor IIF, polypeptide 2 |
| chr5_+_145020910 | 0.37 |
ENSMUST00000124379.3
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr8_-_13939964 | 0.36 |
ENSMUST00000209371.2
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr2_+_91376650 | 0.36 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr5_-_5564730 | 0.36 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr16_-_30152708 | 0.35 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr7_-_110443557 | 0.35 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr6_-_13607963 | 0.35 |
ENSMUST00000031554.9
ENSMUST00000149123.3 |
Tmem168
|
transmembrane protein 168 |
| chr9_+_21634779 | 0.35 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr1_-_106687457 | 0.35 |
ENSMUST00000010049.6
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chrX_-_133442596 | 0.34 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr16_+_16688591 | 0.34 |
ENSMUST00000232080.2
|
Top3b
|
topoisomerase (DNA) III beta |
| chrM_+_8603 | 0.34 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chrX_-_93256244 | 0.34 |
ENSMUST00000113922.2
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr19_+_8779903 | 0.34 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr6_-_69282389 | 0.34 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr10_-_115198093 | 0.34 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr17_+_27775613 | 0.34 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr6_+_17749169 | 0.33 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr5_+_145020640 | 0.32 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr9_+_66853343 | 0.32 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chrX_+_138511360 | 0.32 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr2_-_29677634 | 0.32 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr9_-_21671571 | 0.31 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr3_+_90383425 | 0.31 |
ENSMUST00000001042.10
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr1_+_160898283 | 0.31 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr10_-_129107354 | 0.31 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr5_+_38233901 | 0.30 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr12_+_85157607 | 0.30 |
ENSMUST00000053811.10
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr9_+_113641615 | 0.30 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr10_+_127257077 | 0.30 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr7_+_126575752 | 0.30 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr10_-_62178453 | 0.29 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr10_+_81554753 | 0.29 |
ENSMUST00000085664.6
|
Zfp433
|
zinc finger protein 433 |
| chr14_+_79086492 | 0.29 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr3_+_88486921 | 0.29 |
ENSMUST00000193934.6
ENSMUST00000192495.6 ENSMUST00000035785.9 |
Ssr2
|
signal sequence receptor, beta |
| chr17_+_27775637 | 0.28 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr7_-_127494750 | 0.28 |
ENSMUST00000033074.8
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr6_-_51443602 | 0.28 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr18_+_47245204 | 0.27 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr16_-_90607251 | 0.27 |
ENSMUST00000140920.2
|
Urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr15_-_8740218 | 0.27 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr9_+_74959259 | 0.27 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr15_+_6673167 | 0.27 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr18_+_31742565 | 0.27 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr6_-_115014777 | 0.27 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr9_-_22028370 | 0.26 |
ENSMUST00000213233.2
|
Elof1
|
ELF1 homolog, elongation factor 1 |
| chr5_-_124233812 | 0.26 |
ENSMUST00000031354.11
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr9_+_64188857 | 0.25 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr15_-_37008011 | 0.25 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr9_+_21437440 | 0.25 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr8_-_106692668 | 0.24 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr5_+_30824121 | 0.24 |
ENSMUST00000144742.6
ENSMUST00000149759.2 ENSMUST00000199320.5 |
Cenpa
|
centromere protein A |
| chr17_+_28911529 | 0.24 |
ENSMUST00000114752.3
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr4_+_3940747 | 0.24 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr18_+_37827413 | 0.24 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr8_-_85696040 | 0.24 |
ENSMUST00000214133.2
ENSMUST00000147812.8 |
Gm49661
Rnaseh2a
|
predicted gene, 49661 ribonuclease H2, large subunit |
| chr12_+_65272287 | 0.24 |
ENSMUST00000046331.5
ENSMUST00000221658.2 |
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr10_-_115197775 | 0.24 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr9_-_53521585 | 0.24 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr16_+_16688692 | 0.23 |
ENSMUST00000232547.2
|
Top3b
|
topoisomerase (DNA) III beta |
| chr8_-_85573489 | 0.23 |
ENSMUST00000003912.7
|
Calr
|
calreticulin |
| chr2_-_84481101 | 0.23 |
ENSMUST00000111691.2
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr12_+_55883101 | 0.23 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr5_-_146731810 | 0.23 |
ENSMUST00000085614.6
|
Usp12
|
ubiquitin specific peptidase 12 |
| chr4_+_102446883 | 0.23 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_37249247 | 0.22 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr9_-_119170456 | 0.22 |
ENSMUST00000139870.2
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr2_-_5838489 | 0.22 |
ENSMUST00000128467.4
|
Cdc123
|
cell division cycle 123 |
| chr3_+_88486908 | 0.22 |
ENSMUST00000195014.6
|
Ssr2
|
signal sequence receptor, beta |
| chr6_-_129449739 | 0.22 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr2_+_120807498 | 0.22 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr2_+_112323271 | 0.22 |
ENSMUST00000003705.12
|
Aven
|
apoptosis, caspase activation inhibitor |
| chr1_-_93373145 | 0.21 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr19_+_43741550 | 0.21 |
ENSMUST00000153295.2
|
Cutc
|
cutC copper transporter |
| chr11_-_30218167 | 0.21 |
ENSMUST00000006629.14
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr2_-_84255602 | 0.21 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr4_-_43710231 | 0.20 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr7_+_140521450 | 0.20 |
ENSMUST00000164580.3
ENSMUST00000079403.11 |
Pgghg
|
protein glucosylgalactosylhydroxylysine glucosidase |
| chr11_+_106642052 | 0.20 |
ENSMUST00000147326.9
ENSMUST00000182896.8 ENSMUST00000182908.8 ENSMUST00000086353.11 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr11_+_109434519 | 0.20 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr4_+_5724305 | 0.20 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr2_+_85809620 | 0.20 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr17_+_28226319 | 0.19 |
ENSMUST00000139173.3
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr1_-_80255156 | 0.19 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr16_+_16688605 | 0.19 |
ENSMUST00000232581.2
ENSMUST00000231812.2 ENSMUST00000232017.2 ENSMUST00000023465.16 ENSMUST00000232231.2 ENSMUST00000232200.2 |
Top3b
|
topoisomerase (DNA) III beta |
| chr12_+_79344056 | 0.18 |
ENSMUST00000171210.3
|
Rad51b
|
RAD51 paralog B |
| chr12_+_80690985 | 0.18 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr12_+_10440755 | 0.18 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr15_-_100322089 | 0.18 |
ENSMUST00000154331.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr18_-_39620115 | 0.18 |
ENSMUST00000097592.9
ENSMUST00000115571.8 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr9_-_59260713 | 0.18 |
ENSMUST00000026265.8
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr9_+_35334878 | 0.18 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr11_-_59054521 | 0.17 |
ENSMUST00000137433.2
ENSMUST00000054523.6 |
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr15_+_102234802 | 0.17 |
ENSMUST00000169637.8
|
Pfdn5
|
prefoldin 5 |
| chr9_-_15212849 | 0.17 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr5_+_115417725 | 0.17 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr2_+_174292471 | 0.17 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr7_+_107411470 | 0.17 |
ENSMUST00000208563.3
ENSMUST00000214253.2 |
Olfr467
|
olfactory receptor 467 |
| chr9_-_8134295 | 0.17 |
ENSMUST00000037397.8
|
Cep126
|
centrosomal protein 126 |
| chr1_+_87983099 | 0.16 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr13_-_43634695 | 0.16 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr12_-_85335193 | 0.16 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr4_+_43493344 | 0.16 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr3_-_105861497 | 0.16 |
ENSMUST00000199311.2
|
Atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr17_+_37274714 | 0.16 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr15_-_100322934 | 0.16 |
ENSMUST00000123461.8
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr2_+_109721189 | 0.16 |
ENSMUST00000028583.8
|
Lin7c
|
lin-7 homolog C (C. elegans) |
| chr8_+_124204598 | 0.16 |
ENSMUST00000001520.13
|
Afg3l1
|
AFG3-like AAA ATPase 1 |
| chr11_-_79418500 | 0.16 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr1_+_87983189 | 0.15 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_-_84020382 | 0.15 |
ENSMUST00000018795.13
|
Tada2a
|
transcriptional adaptor 2A |
| chr2_-_85593110 | 0.15 |
ENSMUST00000214958.2
|
Olfr1012
|
olfactory receptor 1012 |
| chr6_-_129655868 | 0.15 |
ENSMUST00000118447.2
ENSMUST00000169545.8 ENSMUST00000032270.13 ENSMUST00000032271.13 |
Klrc1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr17_-_15718542 | 0.15 |
ENSMUST00000014913.11
ENSMUST00000231341.2 |
Psmb1
|
proteasome (prosome, macropain) subunit, beta type 1 |
| chr14_-_101846551 | 0.15 |
ENSMUST00000100340.4
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr6_+_67873135 | 0.15 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr10_+_128067964 | 0.15 |
ENSMUST00000125289.8
ENSMUST00000105242.8 |
Timeless
|
timeless circadian clock 1 |
| chr14_-_55822696 | 0.14 |
ENSMUST00000022828.9
|
Emc9
|
ER membrane protein complex subunit 9 |
| chr14_+_26722319 | 0.14 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr9_+_106699073 | 0.14 |
ENSMUST00000159645.8
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr4_-_82768958 | 0.14 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chrX_-_133012600 | 0.14 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr2_-_68302612 | 0.14 |
ENSMUST00000102715.4
|
Stk39
|
serine/threonine kinase 39 |
| chr8_+_71995554 | 0.14 |
ENSMUST00000034272.9
|
Mvb12a
|
multivesicular body subunit 12A |
| chr16_-_38342949 | 0.14 |
ENSMUST00000002925.6
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr11_+_113575208 | 0.14 |
ENSMUST00000042227.15
ENSMUST00000123466.2 ENSMUST00000106621.4 |
D11Wsu47e
|
DNA segment, Chr 11, Wayne State University 47, expressed |
| chr3_+_66893280 | 0.14 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr7_-_99994257 | 0.14 |
ENSMUST00000207634.2
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr7_+_132212349 | 0.14 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr12_-_104964936 | 0.14 |
ENSMUST00000109927.2
ENSMUST00000095439.11 |
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr18_-_32044877 | 0.14 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr12_-_65113457 | 0.13 |
ENSMUST00000221608.2
|
Fkbp3
|
FK506 binding protein 3 |
| chr5_-_138617952 | 0.13 |
ENSMUST00000063262.11
ENSMUST00000085852.11 ENSMUST00000110905.9 |
Zfp68
|
zinc finger protein 68 |
| chr12_+_65272495 | 0.13 |
ENSMUST00000221980.2
|
Wdr20rt
|
WD repeat domain 20, retrogene |
| chr17_+_56312672 | 0.13 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr11_+_100978103 | 0.13 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prrx1_Isx_Prrxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0018931 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.4 | 1.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.4 | 1.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.3 | 0.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 0.8 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 1.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 1.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 2.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 3.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 1.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.4 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.4 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 2.0 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.7 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.3 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.6 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.6 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.2 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.4 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.2 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 0.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0061341 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.2 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.3 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 1.0 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 1.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.3 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0071459 | kinetochore assembly(GO:0051382) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:0009216 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0009441 | fatty acid alpha-oxidation(GO:0001561) glycolate metabolic process(GO:0009441) |
| 0.0 | 0.3 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 1.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.3 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.3 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 4.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.9 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 3.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.3 | 1.0 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.3 | 3.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.6 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.3 | 0.8 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 1.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.7 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.3 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.3 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.4 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.2 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.4 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.2 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 2.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0051525 | mitogen-activated protein kinase p38 binding(GO:0048273) NFAT protein binding(GO:0051525) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |