Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Ptf1a
Z-value: 0.65
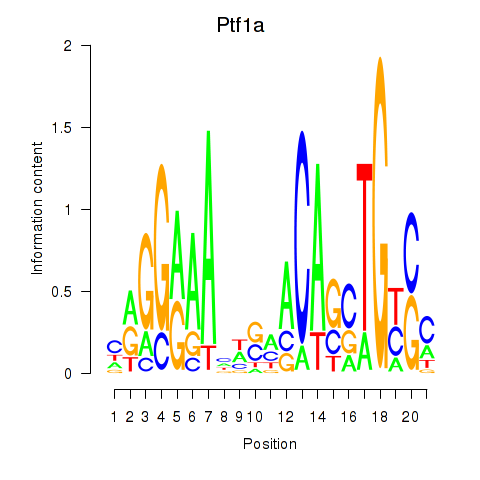
Transcription factors associated with Ptf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ptf1a
|
ENSMUSG00000026735.3 | pancreas specific transcription factor, 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ptf1a | mm39_v1_chr2_+_19450443_19450474 | -0.96 | 1.0e-02 | Click! |
Activity profile of Ptf1a motif
Sorted Z-values of Ptf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_78356523 | 0.41 |
ENSMUST00000001126.4
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr2_+_31360219 | 0.40 |
ENSMUST00000102840.5
|
Ass1
|
argininosuccinate synthetase 1 |
| chr2_+_164790139 | 0.36 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr10_+_75768964 | 0.28 |
ENSMUST00000219839.2
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr11_+_49138278 | 0.21 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr4_+_129083553 | 0.17 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr11_+_40624763 | 0.17 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr2_+_164611812 | 0.16 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr9_+_59563838 | 0.16 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chr6_-_115785695 | 0.15 |
ENSMUST00000081840.6
|
Rpl32
|
ribosomal protein L32 |
| chr10_+_42554888 | 0.15 |
ENSMUST00000040718.6
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr4_-_133615075 | 0.15 |
ENSMUST00000003741.16
ENSMUST00000105894.11 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_-_109502243 | 0.13 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chrX_-_162859429 | 0.13 |
ENSMUST00000134272.2
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B |
| chr7_-_27373939 | 0.13 |
ENSMUST00000138243.2
|
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr8_-_32499556 | 0.13 |
ENSMUST00000207470.3
ENSMUST00000207417.3 ENSMUST00000238826.2 |
Nrg1
|
neuregulin 1 |
| chr5_+_31205971 | 0.12 |
ENSMUST00000013766.13
ENSMUST00000201773.4 ENSMUST00000200748.4 ENSMUST00000201136.2 |
Atraid
|
all-trans retinoic acid induced differentiation factor |
| chr9_+_59563872 | 0.12 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr17_+_8144822 | 0.11 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr8_-_32499513 | 0.11 |
ENSMUST00000208205.3
|
Nrg1
|
neuregulin 1 |
| chr7_-_27374017 | 0.11 |
ENSMUST00000036453.14
ENSMUST00000108341.2 |
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr8_-_106022676 | 0.11 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr8_+_82069177 | 0.11 |
ENSMUST00000213285.2
ENSMUST00000217122.2 ENSMUST00000215332.2 |
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr11_-_6394385 | 0.11 |
ENSMUST00000109737.9
|
H2az2
|
H2A.Z histone variant 2 |
| chr15_+_99568208 | 0.10 |
ENSMUST00000023758.9
|
Asic1
|
acid-sensing (proton-gated) ion channel 1 |
| chr5_+_143450329 | 0.10 |
ENSMUST00000045593.12
|
Daglb
|
diacylglycerol lipase, beta |
| chr19_+_40648182 | 0.10 |
ENSMUST00000112231.9
ENSMUST00000127828.8 |
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr11_+_48691175 | 0.10 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr6_-_5256225 | 0.10 |
ENSMUST00000125686.8
|
Pon3
|
paraoxonase 3 |
| chr10_-_126737185 | 0.10 |
ENSMUST00000168520.3
ENSMUST00000026504.13 |
Atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr11_-_6394352 | 0.09 |
ENSMUST00000093346.6
|
H2az2
|
H2A.Z histone variant 2 |
| chr9_+_106354463 | 0.09 |
ENSMUST00000047721.10
|
Rrp9
|
ribosomal RNA processing 9, U3 small nucleolar RNA binding protein |
| chr19_+_32463151 | 0.09 |
ENSMUST00000025827.10
|
Minpp1
|
multiple inositol polyphosphate histidine phosphatase 1 |
| chr11_+_78215026 | 0.09 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr12_-_31401432 | 0.09 |
ENSMUST00000110857.5
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr6_+_8209216 | 0.08 |
ENSMUST00000040017.8
|
Mios
|
meiosis regulator for oocyte development |
| chr8_+_95584078 | 0.08 |
ENSMUST00000109521.4
|
Polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr3_-_75464066 | 0.08 |
ENSMUST00000162138.2
ENSMUST00000029424.12 ENSMUST00000161137.8 |
Pdcd10
|
programmed cell death 10 |
| chr13_+_19528728 | 0.08 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr11_-_90281721 | 0.08 |
ENSMUST00000004051.8
|
Hlf
|
hepatic leukemia factor |
| chr17_-_28736483 | 0.08 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr11_-_97242842 | 0.07 |
ENSMUST00000093942.5
|
Gpr179
|
G protein-coupled receptor 179 |
| chr14_-_51295099 | 0.07 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr16_-_21980200 | 0.07 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_+_180367056 | 0.07 |
ENSMUST00000094218.4
|
Slc17a9
|
solute carrier family 17, member 9 |
| chr7_+_79939747 | 0.07 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr18_-_80484066 | 0.07 |
ENSMUST00000161003.2
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr7_+_103978678 | 0.07 |
ENSMUST00000180136.2
ENSMUST00000106847.9 |
Trim34b
|
tripartite motif-containing 34B |
| chr1_+_21021889 | 0.07 |
ENSMUST00000038447.6
|
Efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr17_+_29899420 | 0.06 |
ENSMUST00000130052.9
|
Cmtr1
|
cap methyltransferase 1 |
| chr19_-_44057800 | 0.06 |
ENSMUST00000170801.8
|
Erlin1
|
ER lipid raft associated 1 |
| chr11_-_118246566 | 0.06 |
ENSMUST00000155707.3
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr11_+_40624466 | 0.06 |
ENSMUST00000020578.11
|
Nudcd2
|
NudC domain containing 2 |
| chr19_-_9876745 | 0.06 |
ENSMUST00000237725.2
|
Incenp
|
inner centromere protein |
| chr12_+_83572774 | 0.06 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr2_-_34851561 | 0.05 |
ENSMUST00000028234.12
|
Traf1
|
TNF receptor-associated factor 1 |
| chr18_-_80484003 | 0.05 |
ENSMUST00000160434.3
|
Ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr9_-_108474757 | 0.05 |
ENSMUST00000193621.2
ENSMUST00000006853.11 |
P4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr4_-_155095441 | 0.04 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr9_-_108183162 | 0.04 |
ENSMUST00000044725.9
|
Tcta
|
T cell leukemia translocation altered gene |
| chr7_-_97848688 | 0.04 |
ENSMUST00000098278.4
|
B3gnt6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr11_-_102076028 | 0.04 |
ENSMUST00000107156.9
ENSMUST00000021297.6 |
Lsm12
|
LSM12 homolog |
| chr11_+_66847446 | 0.04 |
ENSMUST00000211300.2
ENSMUST00000150220.2 |
Tmem238l
|
transmembrane protein 238 like |
| chr8_+_95584146 | 0.04 |
ENSMUST00000211939.2
ENSMUST00000212124.2 |
Polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr12_-_114451189 | 0.03 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr1_+_36730530 | 0.03 |
ENSMUST00000081180.7
ENSMUST00000193210.6 ENSMUST00000195151.6 |
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr18_+_34464185 | 0.03 |
ENSMUST00000072576.10
ENSMUST00000119329.8 |
Srp19
|
signal recognition particle 19 |
| chr19_+_9960010 | 0.03 |
ENSMUST00000025563.8
ENSMUST00000235407.2 ENSMUST00000237465.2 |
Fth1
|
ferritin heavy polypeptide 1 |
| chr17_+_25527616 | 0.03 |
ENSMUST00000015267.5
|
Prss28
|
protease, serine 28 |
| chr2_-_167852538 | 0.03 |
ENSMUST00000099073.3
|
Ripor3
|
RIPOR family member 3 |
| chr3_-_83749036 | 0.03 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr6_+_65567373 | 0.03 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr7_-_141710850 | 0.03 |
ENSMUST00000209599.2
|
Gm29735
|
predicted gene, 29735 |
| chr11_+_82802079 | 0.03 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_+_25390762 | 0.03 |
ENSMUST00000015239.10
|
Fbxw5
|
F-box and WD-40 domain protein 5 |
| chr12_+_78738425 | 0.03 |
ENSMUST00000218697.2
ENSMUST00000219551.2 ENSMUST00000220396.2 |
Fam71d
|
family with sequence similarity 71, member D |
| chr19_+_16413126 | 0.03 |
ENSMUST00000025602.4
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chr12_-_115276219 | 0.03 |
ENSMUST00000103529.4
|
Ighv1-58
|
immunoglobulin heavy variable 1-58 |
| chr5_-_121025645 | 0.03 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr11_+_120653613 | 0.03 |
ENSMUST00000105046.4
|
Hmga1b
|
high mobility group AT-hook 1B |
| chr9_+_59614877 | 0.03 |
ENSMUST00000128944.8
ENSMUST00000098661.10 |
Gramd2
|
GRAM domain containing 2 |
| chr19_-_13124107 | 0.03 |
ENSMUST00000078299.5
|
Olfr1459
|
olfactory receptor 1459 |
| chr9_+_122850378 | 0.03 |
ENSMUST00000084733.7
ENSMUST00000213514.2 ENSMUST00000217036.2 |
Tmem42
Gm49323
|
transmembrane protein 42 predicted gene, 49323 |
| chr8_-_79975199 | 0.02 |
ENSMUST00000034109.6
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr14_-_24054927 | 0.02 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr9_-_24607554 | 0.02 |
ENSMUST00000133010.4
|
Dpy19l2
|
dpy-19-like 2 (C. elegans) |
| chr2_-_91780731 | 0.02 |
ENSMUST00000111303.2
|
Dgkz
|
diacylglycerol kinase zeta |
| chr6_+_70549568 | 0.02 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr10_+_129376408 | 0.02 |
ENSMUST00000076575.4
|
Olfr792
|
olfactory receptor 792 |
| chr11_+_98337655 | 0.02 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr14_-_119162736 | 0.02 |
ENSMUST00000004055.10
|
Dzip1
|
DAZ interacting protein 1 |
| chr11_-_96807273 | 0.02 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr6_-_69162381 | 0.02 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr19_-_7218512 | 0.02 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr12_+_112727089 | 0.02 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr7_+_17706049 | 0.02 |
ENSMUST00000094799.3
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr4_+_99952988 | 0.02 |
ENSMUST00000039630.6
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr13_-_35108293 | 0.02 |
ENSMUST00000223834.2
|
Fam217a
|
family with sequence similarity 217, member A |
| chr15_-_76501041 | 0.02 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr2_+_142920386 | 0.02 |
ENSMUST00000028902.3
|
Otor
|
otoraplin |
| chr11_-_58346806 | 0.02 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr18_+_37143758 | 0.02 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chr6_-_69220672 | 0.02 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr11_-_73483834 | 0.02 |
ENSMUST00000215689.2
|
Olfr385
|
olfactory receptor 385 |
| chr9_-_112016834 | 0.02 |
ENSMUST00000111872.9
ENSMUST00000164754.9 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr11_-_94595325 | 0.02 |
ENSMUST00000069852.2
|
Gm11541
|
predicted gene 11541 |
| chr10_+_78870557 | 0.02 |
ENSMUST00000082244.3
|
Olfr57
|
olfactory receptor 57 |
| chr2_+_167922924 | 0.02 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr15_+_81943345 | 0.02 |
ENSMUST00000100396.4
|
4930407I10Rik
|
RIKEN cDNA 4930407I10 gene |
| chr11_-_99556781 | 0.02 |
ENSMUST00000100476.3
|
Krtap4-6
|
keratin associated protein 4-6 |
| chr7_+_28455563 | 0.02 |
ENSMUST00000178767.3
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr16_-_63684477 | 0.02 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr6_+_41118120 | 0.02 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr14_-_63654478 | 0.02 |
ENSMUST00000014597.5
|
Blk
|
B lymphoid kinase |
| chr7_-_141674466 | 0.02 |
ENSMUST00000209890.2
|
Gm45618
|
predicted gene 45618 |
| chr2_+_111610658 | 0.02 |
ENSMUST00000054004.2
|
Olfr1302
|
olfactory receptor 1302 |
| chr2_+_22232314 | 0.02 |
ENSMUST00000044749.14
|
Myo3a
|
myosin IIIA |
| chr11_+_99676017 | 0.01 |
ENSMUST00000105059.4
|
Krtap4-9
|
keratin associated protein 4-9 |
| chr12_+_29988035 | 0.01 |
ENSMUST00000122328.8
ENSMUST00000118321.3 |
Pxdn
|
peroxidasin |
| chr7_+_30157704 | 0.01 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr9_-_106353571 | 0.01 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr7_+_141755098 | 0.01 |
ENSMUST00000187512.2
ENSMUST00000084414.6 |
Krtap5-3
|
keratin associated protein 5-3 |
| chr14_+_50645025 | 0.01 |
ENSMUST00000214853.2
ENSMUST00000214320.2 |
Olfr738
|
olfactory receptor 738 |
| chr6_+_41279199 | 0.01 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr13_-_61012185 | 0.01 |
ENSMUST00000091569.7
|
4930486L24Rik
|
RIKEN cDNA 4930486L24 gene |
| chr12_+_78738288 | 0.01 |
ENSMUST00000077968.5
|
Fam71d
|
family with sequence similarity 71, member D |
| chr14_-_70445086 | 0.01 |
ENSMUST00000022682.6
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr16_-_55103598 | 0.01 |
ENSMUST00000036412.4
|
Zpld1
|
zona pellucida like domain containing 1 |
| chr17_-_26420332 | 0.01 |
ENSMUST00000121959.3
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr14_-_56181993 | 0.01 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr7_-_106341163 | 0.01 |
ENSMUST00000050541.5
|
Olfr697
|
olfactory receptor 697 |
| chr6_+_41331039 | 0.01 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr9_-_44646487 | 0.01 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr2_-_88559941 | 0.01 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr17_-_27732364 | 0.01 |
ENSMUST00000118161.3
|
Grm4
|
glutamate receptor, metabotropic 4 |
| chr1_+_82938007 | 0.01 |
ENSMUST00000223536.2
|
Gm47955
|
predicted gene, 47955 |
| chr8_-_71315902 | 0.01 |
ENSMUST00000212611.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr7_-_142628298 | 0.01 |
ENSMUST00000148715.8
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr17_+_24087030 | 0.01 |
ENSMUST00000024928.4
|
Prss21
|
protease, serine 21 |
| chr4_-_88552379 | 0.01 |
ENSMUST00000156062.2
ENSMUST00000107142.3 ENSMUST00000107143.8 ENSMUST00000053304.10 |
Pramel32
|
PRAME like 32 |
| chr3_+_117368876 | 0.01 |
ENSMUST00000106473.5
|
Plppr5
|
phospholipid phosphatase related 5 |
| chr16_-_63684425 | 0.01 |
ENSMUST00000232049.2
|
Epha3
|
Eph receptor A3 |
| chr7_+_17799889 | 0.01 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr4_-_133930315 | 0.01 |
ENSMUST00000097849.3
|
Zpld2
|
zona pellucida like domain containing 2 |
| chr4_+_118516149 | 0.01 |
ENSMUST00000213189.3
|
Olfr62
|
olfactory receptor 62 |
| chr11_+_98489117 | 0.01 |
ENSMUST00000142268.3
|
Lrrc3c
|
leucine rich repeat containing 3C |
| chr17_-_26420300 | 0.01 |
ENSMUST00000025019.9
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr3_+_117368483 | 0.01 |
ENSMUST00000039564.11
ENSMUST00000238937.2 |
Plppr5
|
phospholipid phosphatase related 5 |
| chr3_-_141637245 | 0.01 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr16_-_29363671 | 0.01 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr6_+_41435846 | 0.01 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr14_-_32907023 | 0.01 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr2_-_93787383 | 0.01 |
ENSMUST00000148314.3
|
Gm13889
|
predicted gene 13889 |
| chr4_+_90111862 | 0.01 |
ENSMUST00000080541.5
|
Zfp352
|
zinc finger protein 352 |
| chr12_-_103641723 | 0.01 |
ENSMUST00000095451.2
|
Serpina16
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
| chr14_-_50536787 | 0.01 |
ENSMUST00000163469.2
|
Olfr733
|
olfactory receptor 733 |
| chr8_-_112417633 | 0.01 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr11_-_99695272 | 0.01 |
ENSMUST00000105056.2
|
Gm11554
|
predicted gene 11554 |
| chr6_-_68784692 | 0.01 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr9_+_36604516 | 0.01 |
ENSMUST00000034620.5
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr7_-_130924021 | 0.01 |
ENSMUST00000046611.9
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr10_-_75768302 | 0.01 |
ENSMUST00000120281.8
ENSMUST00000000924.13 |
Mmp11
|
matrix metallopeptidase 11 |
| chr12_+_37930305 | 0.01 |
ENSMUST00000220990.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr12_-_87283780 | 0.01 |
ENSMUST00000221768.2
|
Noxred1
|
NADP+ dependent oxidoreductase domain containing 1 |
| chrX_+_156601431 | 0.01 |
ENSMUST00000087157.5
|
Klhl34
|
kelch-like 34 |
| chr7_-_106491314 | 0.01 |
ENSMUST00000088687.3
|
Olfr707
|
olfactory receptor 707 |
| chr16_-_44153288 | 0.01 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr7_+_118199375 | 0.01 |
ENSMUST00000121744.9
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr4_-_36056726 | 0.01 |
ENSMUST00000108124.4
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr12_-_114263874 | 0.01 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr1_-_172156828 | 0.01 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chrX_+_100427331 | 0.01 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr11_-_70213348 | 0.01 |
ENSMUST00000019051.3
|
Alox12e
|
arachidonate lipoxygenase, epidermal |
| chr5_+_7354130 | 0.01 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr6_-_69261303 | 0.01 |
ENSMUST00000103349.2
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr7_-_79036222 | 0.01 |
ENSMUST00000205638.2
ENSMUST00000206320.3 ENSMUST00000205442.2 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr9_+_19408046 | 0.01 |
ENSMUST00000064582.6
|
Olfr851
|
olfactory receptor 851 |
| chr6_-_70318164 | 0.01 |
ENSMUST00000103389.3
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr8_+_110827839 | 0.01 |
ENSMUST00000213641.2
|
Tle7
|
TLE family member 7 |
| chr10_-_95253042 | 0.01 |
ENSMUST00000135822.8
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr4_+_85432409 | 0.01 |
ENSMUST00000213098.2
|
Adamtsl1
|
ADAMTS-like 1 |
| chr3_-_80820835 | 0.01 |
ENSMUST00000107743.8
ENSMUST00000029654.15 |
Glrb
|
glycine receptor, beta subunit |
| chr18_+_32055339 | 0.01 |
ENSMUST00000233994.2
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr4_+_118522716 | 0.00 |
ENSMUST00000102666.5
|
Olfr62
|
olfactory receptor 62 |
| chr8_+_84016970 | 0.00 |
ENSMUST00000034146.5
|
Ucp1
|
uncoupling protein 1 (mitochondrial, proton carrier) |
| chr3_+_137983250 | 0.00 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr2_+_4980986 | 0.00 |
ENSMUST00000027978.7
ENSMUST00000195688.2 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr7_-_79036691 | 0.00 |
ENSMUST00000053718.15
ENSMUST00000179243.3 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr2_-_86254018 | 0.00 |
ENSMUST00000105213.5
|
Olfr1062
|
olfactory receptor 1062 |
| chr17_+_21910767 | 0.00 |
ENSMUST00000072133.5
|
Gm10226
|
predicted gene 10226 |
| chr15_-_77283286 | 0.00 |
ENSMUST00000175919.8
ENSMUST00000176074.9 |
Apol7a
|
apolipoprotein L 7a |
| chr9_+_108743687 | 0.00 |
ENSMUST00000026744.6
ENSMUST00000192852.2 |
Tmem89
|
transmembrane protein 89 |
| chr5_+_149363114 | 0.00 |
ENSMUST00000031667.8
|
Tex26
|
testis expressed 26 |
| chr8_+_123920682 | 0.00 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr8_-_120904159 | 0.00 |
ENSMUST00000135567.4
|
Cibar2
|
CBY1 interacting BAR domain containing 2 |
| chr4_+_88692071 | 0.00 |
ENSMUST00000177806.2
|
Gm13290
|
predicted gene 13290 |
| chr6_-_136758716 | 0.00 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr1_+_151074199 | 0.00 |
ENSMUST00000225273.2
|
Gm47995
|
predicted gene, 47995 |
| chr3_+_94391676 | 0.00 |
ENSMUST00000198384.3
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr6_-_41291634 | 0.00 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ptf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.4 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.3 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) methotrexate transport(GO:0051958) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.0 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0052572 | positive regulation of interleukin-18 production(GO:0032741) detection of triacyl bacterial lipopeptide(GO:0042495) response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) response to host(GO:0075136) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.0 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |